Definitions
127 definitions
🧬 Abrogated
🧬 Alternative Splicing
⚓ Anchors (URL Fragments)
📊 ANSI SQL
🧬 AT-hooks
🧪 ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing)
👀 Attention Mechanism
🔧 Autograd
↩️ Backpropagation
🧮 BAF and PBAF Complexes
🔄 Canonical Polyadic (CP) Decomposition
⛓️ Chain Rule
🧬 Chimeric
🧩 Chinese Remainder Theorem
🔬 ChIP-seq (Chromatin Immunoprecipitation Sequencing)
🔄 Chirality
🧪 Chromatin Remodeling
🔍 Chromodomains and Bromodomains
🧬 Cis DNA Regulatory Elements
👁️ Convolutional Neural Network (CNN)
🌐 CORS (Cross-Origin Resource Sharing)
📊 Cross Entropy Loss
🧪 Cytotoxicity
⚖️ Dalton (Da)
🔬 Deconvolution Analysis
🔮 Deep Learning
🦕 Deno
🔐 Diffie-Hellman Key Exchange
🔄 Dimers
🌐 DNS (Domain Name System)
🏠 Domain
📚 Edify
🔀 Elastic Net
🔠 Embedding
🧬 Endogenous
🧬 Epochs
📐 Euler's Number (e)
🔢 Exponent Rules
🎯 Fine-tuning
🔢🔍 Float (Floating-Point Number)
🔬📊 Floating-Point Precision
🧬 Gene Fusion Events
🌐 Generalized CP (GCP) Decomposition
🎨 Generative Adversarial Network (GAN)
🔍✅ Git Triage:
⬇️ Gradient Descent
🧠 Hebbian Theory
🧩 Hi-C (Chromosome Conformation Capture + Sequencing)
🧬 HLA Imputation
🧬 Homologs
🌐 Internet Protocol (IP)
🌐 IP Address (Internet Protocol Address)
🧬 Isoforms
🧪 Kinases
📊 Lasso Regression
🧠 Long Short-Term Memory (LSTM)
📊 Loss Function
🤖 Machine Learning
🔬 Mechanistic Modeling
📊 Mixed-Effects Models
🧬 Moiety
📈 Monotonic
🔄 Monovalent Molecular Glue Degraders
🧩 Mosaicism Detection
🔗 Multi-Layer Perceptron (MLP)
🩸 Myelodysplastic Syndrome (MDS)
🧠 Neural Network
🟢 Node.js
📈 Non-parametric Statistics
🔍⏱️ NP (Nondeterministic Polynomial Time)
🧩🔄 NP-Complete
🏋️♂️🧠 NP-Hard
⚡🎯 Optimizer
🧬 Orthologs
📈 Overfitting
⏱️✓ P (Polynomial Time)
🧬 Paralogs
📊 Parametric Statistics
📊 Pearson Correlation Coefficient
🧬 Pleiotropy
🔄 Pluripotency
🧮 Pohlig-Hellman Algorithm
🚪 Ports
📍 Positional Encoding
⌨️ Prompt Engineering
🧬 PROteolysis TArgeting Chimeras (PROTACs)
🔗 Protocol/Scheme
➗ Quotient Rule
🔄 Recurrent Neural Network (RNN)
🎮 Reinforcement Learning
⚡ ReLU (Rectified Linear Unit)
🔄🚀 Repository Dispatch:
🌐 RESTful API
📈 Ridge Regression
🧬 RNA-seq (RNA sequencing)
🧬 sgRNA (Single Guide RNA)
🔒🧮 SHA-256 Checksum
🔗 Similarity Network Fusion (SNF)
🔓 Small Subgroup Vulnerabilities
📊 Softmax
🧬 Somatic Mutations
🔐💻 SSH (Secure Shell)
🔄 Stereoisomers
🎲 Stochastic
🌐 Subdomain
👨🏫 Supervised Learning
🧪 Svedberg Sedimentation Coefficient
🎯 Targeted Protein Degradation
🤝📬 TCP (Transmission Control Protocol)
📊 Tensor Decomposition
🔢 Tensor Processing Unit (TPU)
🔒 TLS (Transport Layer Security)
✂️ Tokenization
🧬 Transcription Factor (TF) Networks
🔄 Transformer
🧩 Tucker Decomposition
🔄 Ubiquitin Proteasome System (UPS)
🚀📨 UDP (User Datagram Protocol)
🔍 Unsupervised Learning
🔗 URL (Uniform Resource Locator)
🗜️ Variational Autoencoder (VAE)
🗄️ Vector Database
🔌💻 Verilog
🧫🔬 Western Blot/Immunoblot
📝 Word Embedding
🧬 Xenologs
🧬 Zinc Fingers
All Definitions
Filter by tag:
🧬 Abrogated
dictionary
Refers to something that has been abolished, terminated, or suppressed. In molecular biology and immunology, it specifically describes the blocking or suppression of a biological function or process, particularly an immune response. When a gene, protein, or cellular pathway is abrogated, its normal function has been inhibited or eliminated.
🧬 Alternative Splicing
biology
• Definition: A post-transcriptional process where different exons from a pre-mRNA are included in or excluded from the mature mRNA, generating multiple transcript variants from a single gene.
• Major types:
- Exon skipping: Exclusion of an exon from the final transcript
- Mutually exclusive exons: Inclusion of one exon from a set of possible exons
- Alternative 5' splice sites: Use of different 5' splice junctions
- Alternative 3' splice sites: Use of different 3' splice junctions
- Intron retention: Inclusion of an intron in the mature transcript
• Regulatory mechanisms:
- Cis-regulatory elements: Exonic/intronic splicing enhancers or silencers
- Trans-acting factors: SR proteins, hnRNPs, and other splicing regulators
- RNA secondary structure: Affects accessibility of splice sites
- Epigenetic marks: Histone modifications and DNA methylation
- Transcription rate: Kinetic coupling between transcription and splicing
• Biological importance:
- Expands transcriptome and proteome diversity
- Enables tissue-specific and developmental stage-specific gene expression
- Contributes to evolutionary adaptation and species complexity
- Allows rapid cellular responses to environmental changes
• Detection and analysis methods:
- RNA-seq with splice junction analysis
- Exon microarrays
- RT-PCR with -specific primers
- Computational tools: MISO, rMATS, SUPPA, Whippet
• Clinical significance:
- Splicing dysregulation in cancer and neurodegenerative diseases
- Mutations affecting splice sites cause ~15% of human genetic diseases
- Therapeutic approaches targeting splicing (antisense oligonucleotides, small molecules)
- Splicing patterns as diagnostic or prognostic biomarkers
• Major types:
- Exon skipping: Exclusion of an exon from the final transcript
- Mutually exclusive exons: Inclusion of one exon from a set of possible exons
- Alternative 5' splice sites: Use of different 5' splice junctions
- Alternative 3' splice sites: Use of different 3' splice junctions
- Intron retention: Inclusion of an intron in the mature transcript
• Regulatory mechanisms:
- Cis-regulatory elements: Exonic/intronic splicing enhancers or silencers
- Trans-acting factors: SR proteins, hnRNPs, and other splicing regulators
- RNA secondary structure: Affects accessibility of splice sites
- Epigenetic marks: Histone modifications and DNA methylation
- Transcription rate: Kinetic coupling between transcription and splicing
• Biological importance:
- Expands transcriptome and proteome diversity
- Enables tissue-specific and developmental stage-specific gene expression
- Contributes to evolutionary adaptation and species complexity
- Allows rapid cellular responses to environmental changes
• Detection and analysis methods:
- RNA-seq with splice junction analysis
- Exon microarrays
- RT-PCR with -specific primers
- Computational tools: MISO, rMATS, SUPPA, Whippet
• Clinical significance:
- Splicing dysregulation in cancer and neurodegenerative diseases
- Mutations affecting splice sites cause ~15% of human genetic diseases
- Therapeutic approaches targeting splicing (antisense oligonucleotides, small molecules)
- Splicing patterns as diagnostic or prognostic biomarkers
⚓ Anchors (URL Fragments)
web-developmenthtml
The part of a URL that comes after the hash symbol (#) and refers to a specific section or element within a web page. Anchors allow users to jump directly to particular content on a page without scrolling through the entire document.
Anchors are also called fragments, hash fragments, or URL fragments. They are processed by the browser and typically don't get sent to the server as part of the HTTP request.
Anchor Examples:
- `https://example.com/page#section1` - Jumps to element with id="section1"
- `https://example.com/docs#installation` - Jumps to installation section
- `https://example.com/article#conclusion` - Jumps to conclusion
Common Use Cases:
- Table of Contents: Quick navigation to different sections
- Documentation: Linking to specific topics or API methods
- Long Articles: Jumping to particular paragraphs or chapters
- Single Page Applications: Navigation without page reloads
- Form Validation: Scrolling to error fields
Technical Notes:
- Anchors target HTML elements with matching `id` attributes
- They don't trigger new HTTP requests
- Can be used with JavaScript for dynamic behavior
- Useful for SEO and user experience
- Supported by all modern browsers
Anchors are also called fragments, hash fragments, or URL fragments. They are processed by the browser and typically don't get sent to the server as part of the HTTP request.
Anchor Examples:
- `https://example.com/page#section1` - Jumps to element with id="section1"
- `https://example.com/docs#installation` - Jumps to installation section
- `https://example.com/article#conclusion` - Jumps to conclusion
Common Use Cases:
- Table of Contents: Quick navigation to different sections
- Documentation: Linking to specific topics or API methods
- Long Articles: Jumping to particular paragraphs or chapters
- Single Page Applications: Navigation without page reloads
- Form Validation: Scrolling to error fields
Technical Notes:
- Anchors target HTML elements with matching `id` attributes
- They don't trigger new HTTP requests
- Can be used with JavaScript for dynamic behavior
- Useful for SEO and user experience
- Supported by all modern browsers
📊 ANSI SQL
computer science
The American National Standards Institute (ANSI) standard for the Structured Query Language (SQL), which defines a standardized syntax and functionality for relational database management systems. ANSI SQL ensures portability and consistency across different database platforms by establishing common rules for data definition, manipulation, and querying.
Key features of ANSI SQL include:
Data Definition Language (DDL): CREATE, ALTER, DROP statements for database schema
Data Manipulation Language (DML): SELECT, INSERT, UPDATE, DELETE for data operations
Data Control Language (DCL): GRANT, REVOKE for access control
Transaction Control: COMMIT, ROLLBACK for data integrity
Standard Functions: Aggregate functions (SUM, COUNT, AVG), string functions, date functions
Join Operations: INNER, LEFT, RIGHT, FULL OUTER joins
Subqueries and CTEs: Common Table Expressions for complex queries
ANSI SQL has evolved through several versions (SQL-86, SQL-89, SQL-92, SQL:1999, SQL:2003, SQL:2006, SQL:2008, SQL:2011, SQL:2016) with each adding new features like window functions, JSON support, and advanced analytics. While most database systems support core ANSI SQL, they often include proprietary extensions for enhanced functionality.
Key features of ANSI SQL include:
Data Definition Language (DDL): CREATE, ALTER, DROP statements for database schema
Data Manipulation Language (DML): SELECT, INSERT, UPDATE, DELETE for data operations
Data Control Language (DCL): GRANT, REVOKE for access control
Transaction Control: COMMIT, ROLLBACK for data integrity
Standard Functions: Aggregate functions (SUM, COUNT, AVG), string functions, date functions
Join Operations: INNER, LEFT, RIGHT, FULL OUTER joins
Subqueries and CTEs: Common Table Expressions for complex queries
ANSI SQL has evolved through several versions (SQL-86, SQL-89, SQL-92, SQL:1999, SQL:2003, SQL:2006, SQL:2008, SQL:2011, SQL:2016) with each adding new features like window functions, JSON support, and advanced analytics. While most database systems support core ANSI SQL, they often include proprietary extensions for enhanced functionality.
🧬 AT-hooks
biology
• Definition: Small DNA-binding motifs that recognize and bind to the minor groove of AT-rich DNA sequences.
• Structure:
- Characterized by a conserved core sequence of Arg-Gly-Arg-Pro (RGRP)
- Typically flanked by positively charged amino acids (lysine or arginine)
- Intrinsically disordered regions that adopt a defined structure upon DNA binding
- Often present in multiple copies within a single protein
• Biological functions:
- Chromatin architecture: Contributing to higher-order chromatin structure organization
- Transcriptional regulation: Modulating gene expression by altering DNA accessibility
- Enhancer binding: Recognizing AT-rich enhancer elements
- Protein-protein interactions: Serving as platforms for recruiting other regulatory proteins
• Notable AT-hook containing proteins:
- HMGA family (High Mobility Group A): Non-histone chromosomal proteins involved in gene regulation
- PRC1 complex components: Involved in Polycomb-mediated gene silencing
- Various transcription factors: Including SOX2, OCT4, and other developmental regulators
• Clinical relevance:
- Cancer: Aberrant expression of AT-hook proteins (especially HMGA) in various tumors
- Developmental disorders: Mutations in AT-hook proteins linked to growth abnormalities
- Potential therapeutic targets: Emerging strategies targeting AT-hook/DNA interactions
• Structure:
- Characterized by a conserved core sequence of Arg-Gly-Arg-Pro (RGRP)
- Typically flanked by positively charged amino acids (lysine or arginine)
- Intrinsically disordered regions that adopt a defined structure upon DNA binding
- Often present in multiple copies within a single protein
• Biological functions:
- Chromatin architecture: Contributing to higher-order chromatin structure organization
- Transcriptional regulation: Modulating gene expression by altering DNA accessibility
- Enhancer binding: Recognizing AT-rich enhancer elements
- Protein-protein interactions: Serving as platforms for recruiting other regulatory proteins
• Notable AT-hook containing proteins:
- HMGA family (High Mobility Group A): Non-histone chromosomal proteins involved in gene regulation
- PRC1 complex components: Involved in Polycomb-mediated gene silencing
- Various transcription factors: Including SOX2, OCT4, and other developmental regulators
• Clinical relevance:
- Cancer: Aberrant expression of AT-hook proteins (especially HMGA) in various tumors
- Developmental disorders: Mutations in AT-hook proteins linked to growth abnormalities
- Potential therapeutic targets: Emerging strategies targeting AT-hook/DNA interactions
🧪 ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing)
biologylab-techniques
• Purpose: Identifies open or accessible regions of chromatin — places where DNA is not tightly wrapped around histones and is more likely to be active.
• Data type: Peaks indicating open chromatin regions.
• Used for: Inferring regulatory regions like enhancers, promoters, and transcription factor binding sites.
• Data type: Peaks indicating open chromatin regions.
• Used for: Inferring regulatory regions like enhancers, promoters, and transcription factor binding sites.
👀 Attention Mechanism
machine-learning
A breakthrough technique that allows to focus on relevant parts of input data, similar to human attention. By learning which parts of the input are important for each output, it dramatically improves performance in tasks like translation and image captioning. This mechanism is a key component of architectures.
🔧 Autograd
mathmachine-learningcomputer science
Automatic differentiation (autograd) is a computational technique that automatically calculates derivatives of functions defined by computer programs. Unlike symbolic differentiation (which manipulates mathematical expressions) or numerical differentiation (which approximates derivatives using finite differences), autograd computes exact derivatives efficiently by applying the systematically during program execution.
In , autograd is fundamental to training through gradient-based optimization. It enables frameworks like PyTorch, TensorFlow, and JAX to automatically compute gradients of with respect to model parameters, eliminating the need for manual derivative calculations. This automation is crucial for , where models may have millions or billions of parameters.
Autograd works by tracking operations performed on tensors and building a computational graph that records how outputs depend on inputs. During the backward pass, it traverses this graph in reverse order, applying the to compute gradients efficiently. This process, combined with , enables the training of complex neural architectures that would be impractical to differentiate manually.
Modern autograd systems support both forward-mode and reverse-mode automatic differentiation, with reverse-mode (used in ) being particularly efficient for functions with many inputs and few outputs, which is typical in scenarios.
Simple Examples:
1. Strong Positive Correlation (r ≈ 0.9): Height and weight in a population. As height increases, weight tends to increase proportionally.
2. Moderate Positive Correlation (r ≈ 0.4): Study hours and test scores. More study time generally leads to better scores, but other factors also influence performance.
3. No Correlation (r ≈ 0): Shoe size and intelligence. These variables have no meaningful linear relationship.
4. Moderate Negative Correlation (r ≈ -0.4): Age of a car and its resale value. Older cars typically have lower resale values, though condition and other factors matter.
5. Strong Negative Correlation (r ≈ -0.8): Outdoor temperature and home heating usage. As temperature drops, heating usage increases substantially.
In , autograd is fundamental to training through gradient-based optimization. It enables frameworks like PyTorch, TensorFlow, and JAX to automatically compute gradients of with respect to model parameters, eliminating the need for manual derivative calculations. This automation is crucial for , where models may have millions or billions of parameters.
Autograd works by tracking operations performed on tensors and building a computational graph that records how outputs depend on inputs. During the backward pass, it traverses this graph in reverse order, applying the to compute gradients efficiently. This process, combined with , enables the training of complex neural architectures that would be impractical to differentiate manually.
Modern autograd systems support both forward-mode and reverse-mode automatic differentiation, with reverse-mode (used in ) being particularly efficient for functions with many inputs and few outputs, which is typical in scenarios.
Simple Examples:
1. Strong Positive Correlation (r ≈ 0.9): Height and weight in a population. As height increases, weight tends to increase proportionally.
2. Moderate Positive Correlation (r ≈ 0.4): Study hours and test scores. More study time generally leads to better scores, but other factors also influence performance.
3. No Correlation (r ≈ 0): Shoe size and intelligence. These variables have no meaningful linear relationship.
4. Moderate Negative Correlation (r ≈ -0.4): Age of a car and its resale value. Older cars typically have lower resale values, though condition and other factors matter.
5. Strong Negative Correlation (r ≈ -0.8): Outdoor temperature and home heating usage. As temperature drops, heating usage increases substantially.
↩️ Backpropagation
machine-learning
The core learning algorithm for that calculates how much each neuron contributed to the error, then adjusts weights backwards through the network. It's like tracing back through a chain of decisions to identify and correct mistakes, enabling efficient network training.
[The Most Important Algorithm in ](https://www.youtube.com/watch?v=SmZmBKc7Lrs&ab_channel=ArtemKirsanov)
[The Most Important Algorithm in ](https://www.youtube.com/watch?v=SmZmBKc7Lrs&ab_channel=ArtemKirsanov)
🧮 BAF and PBAF Complexes
biology
• Definition: ATP-dependent complexes that alter nucleosome positioning to regulate DNA accessibility and gene expression.
• Composition: Both are variants of the SWI/SNF (SWItch/Sucrose Non-Fermentable) complex family with:
- Shared core subunits: BRG1/BRM (ATPase), BAF155, BAF170, BAF60
- Distinctive subunits: PBAF contains PBRM1, BAF200, and BRD7; BAF contains BAF250A/B (ARID1A/B)
• Function:
- Nucleosome repositioning and ejection to control DNA accessibility
- Transcriptional regulation through interaction with transcription factors
- Chromatin boundary establishment and maintenance
- Cell lineage determination and differentiation
• Disease associations:
- Cancer: Mutations in BAF/PBAF subunits occur in ~20% of human cancers
- Neurodevelopmental disorders: Mutations linked to autism, intellectual disability
- Coffin-Siris and Nicolaides-Baraitser syndromes: Caused by mutations in BAF complex genes
• Therapeutic relevance: Emerging targets for cancer therapy through synthetic lethality approaches and epigenetic modulation.
• Composition: Both are variants of the SWI/SNF (SWItch/Sucrose Non-Fermentable) complex family with:
- Shared core subunits: BRG1/BRM (ATPase), BAF155, BAF170, BAF60
- Distinctive subunits: PBAF contains PBRM1, BAF200, and BRD7; BAF contains BAF250A/B (ARID1A/B)
• Function:
- Nucleosome repositioning and ejection to control DNA accessibility
- Transcriptional regulation through interaction with transcription factors
- Chromatin boundary establishment and maintenance
- Cell lineage determination and differentiation
• Disease associations:
- Cancer: Mutations in BAF/PBAF subunits occur in ~20% of human cancers
- Neurodevelopmental disorders: Mutations linked to autism, intellectual disability
- Coffin-Siris and Nicolaides-Baraitser syndromes: Caused by mutations in BAF complex genes
• Therapeutic relevance: Emerging targets for cancer therapy through synthetic lethality approaches and epigenetic modulation.
🔄 Canonical Polyadic (CP) Decomposition
mathmachine-learning
Also known as CANDECOMP/PARAFAC decomposition, CP breaks down a tensor into a sum of rank-one tensors (outer products of vectors). This decomposition provides a highly interpretable representation where each component represents a distinct pattern or factor in the data.
CP decomposition serves as a powerful tool for discovering latent factors in multi-way data, with applications in chemometrics (analyzing chemical measurements), neuroscience (identifying functional networks), and recommendation systems (capturing user-item-context interactions).
CP decomposition serves as a powerful tool for discovering latent factors in multi-way data, with applications in chemometrics (analyzing chemical measurements), neuroscience (identifying functional networks), and recommendation systems (capturing user-item-context interactions).
⛓️ Chain Rule
mathmachine-learning
A fundamental rule in calculus for finding the derivative of composite functions. The chain rule states that if you have a composite function f(g(x)), then its derivative is the derivative of the outer function evaluated at the inner function, multiplied by the derivative of the inner function.
Mathematically expressed as:
Or in Leibniz notation:
The chain rule is essential for differentiating complex functions and is widely used in calculus, physics, engineering, and (particularly in algorithms for ). Common applications include finding derivatives of exponential functions, trigonometric functions with inner functions, and nested polynomial expressions.
Mathematically expressed as:
Or in Leibniz notation:
The chain rule is essential for differentiating complex functions and is widely used in calculus, physics, engineering, and (particularly in algorithms for ). Common applications include finding derivatives of exponential functions, trigonometric functions with inner functions, and nested polynomial expressions.
🧬 Chimeric
dictionarybiology
Refers to something that is made from parts originating from different sources—combined into a single entity.
🧩 Chinese Remainder Theorem
math
A fundamental result in number theory that provides a solution to systems of simultaneous linear congruences with coprime moduli. The theorem states that if one has several congruence equations, a unique solution exists modulo the product of the moduli, provided that the moduli are pairwise coprime.
Formally, if n₁, n₂, ..., nₖ are pairwise coprime positive integers and a₁, a₂, ..., aₖ are any integers, then the system of congruences x ≡ a₁ (mod n₁), x ≡ a₂ (mod n₂), ..., x ≡ aₖ (mod nₖ) has a unique solution modulo N = n₁ × n₂ × ... × nₖ.
The theorem has applications in various fields including cryptography (RSA algorithm), coding theory, and computer science (particularly in distributed computing and for creating efficient algorithms). It also has historical significance, originating in ancient Chinese mathematics as early as the 3rd century CE in the mathematical text "Sunzi Suanjing."
Video explanation:
Chinese Remainder Theorem - A comprehensive explanation of the Chinese Remainder Theorem, its proof, and applications.
Formally, if n₁, n₂, ..., nₖ are pairwise coprime positive integers and a₁, a₂, ..., aₖ are any integers, then the system of congruences x ≡ a₁ (mod n₁), x ≡ a₂ (mod n₂), ..., x ≡ aₖ (mod nₖ) has a unique solution modulo N = n₁ × n₂ × ... × nₖ.
The theorem has applications in various fields including cryptography (RSA algorithm), coding theory, and computer science (particularly in distributed computing and for creating efficient algorithms). It also has historical significance, originating in ancient Chinese mathematics as early as the 3rd century CE in the mathematical text "Sunzi Suanjing."
Video explanation:
Chinese Remainder Theorem - A comprehensive explanation of the Chinese Remainder Theorem, its proof, and applications.
🔬 ChIP-seq (Chromatin Immunoprecipitation Sequencing)
biologylab-techniques
• Purpose: Maps protein-DNA interactions, such as where transcription factors or modified histones bind DNA.
• Data type: Enrichment peaks showing binding locations.
• Used for: Studying gene regulation, histone modifications, and epigenetic changes.
• Data type: Enrichment peaks showing binding locations.
• Used for: Studying gene regulation, histone modifications, and epigenetic changes.
🔄 Chirality
biology
• Definition: The geometric property where a molecule cannot be superimposed on its mirror image, similar to how left and right hands are non-superimposable mirror images of each other.
• Key concepts:
- Chiral center: Typically a carbon atom bonded to four different groups
- Enantiomers: Mirror-image forms of a chiral molecule
- Optical activity: Chiral molecules rotate plane-polarized light
- Racemic mixture: Equal mixture of both enantiomers
• Biological importance:
- Enzyme specificity: Most enzymes interact with only one enantiomer of a substrate
- Drug efficacy and safety: Different enantiomers can have dramatically different biological effects
- Protein structure: All natural amino acids (except glycine) are chiral
- Nucleic acid structure: The sugar component in DNA and RNA is chiral
• Nomenclature systems:
- R/S system: Based on Cahn-Ingold-Prelog priority rules
- D/L system: Based on the configuration of glyceraldehyde
- (+)/(-) system: Based on the direction of rotation of plane-polarized light
• Applications in bioinformatics and computational chemistry:
- Molecular modeling: Accurate representation of 3D molecular structures
- Drug discovery: Virtual screening of specific enantiomers
- Protein-ligand interactions: Predicting binding affinities of chiral molecules
- Cheminformatics: Algorithms for detecting and representing chirality
• Key concepts:
- Chiral center: Typically a carbon atom bonded to four different groups
- Enantiomers: Mirror-image forms of a chiral molecule
- Optical activity: Chiral molecules rotate plane-polarized light
- Racemic mixture: Equal mixture of both enantiomers
• Biological importance:
- Enzyme specificity: Most enzymes interact with only one enantiomer of a substrate
- Drug efficacy and safety: Different enantiomers can have dramatically different biological effects
- Protein structure: All natural amino acids (except glycine) are chiral
- Nucleic acid structure: The sugar component in DNA and RNA is chiral
• Nomenclature systems:
- R/S system: Based on Cahn-Ingold-Prelog priority rules
- D/L system: Based on the configuration of glyceraldehyde
- (+)/(-) system: Based on the direction of rotation of plane-polarized light
• Applications in bioinformatics and computational chemistry:
- Molecular modeling: Accurate representation of 3D molecular structures
- Drug discovery: Virtual screening of specific enantiomers
- Protein-ligand interactions: Predicting binding affinities of chiral molecules
- Cheminformatics: Algorithms for detecting and representing chirality
🧪 Chromatin Remodeling
biology
• Definition: The dynamic process by which specialized protein complexes alter chromatin structure to regulate DNA accessibility for transcription, replication, repair, and recombination.
• Mechanisms:
- Nucleosome sliding (moving histone proteins): Repositioning nucleosomes along DNA without disrupting histone-DNA contacts
- Histone eviction/replacement: Removing or exchanging histones to alter chromatin composition
- Histone modification: Adding or removing chemical groups that affect chromatin compaction
- ATP-dependent remodeling: Using energy from ATP hydrolysis to physically restructure chromatin
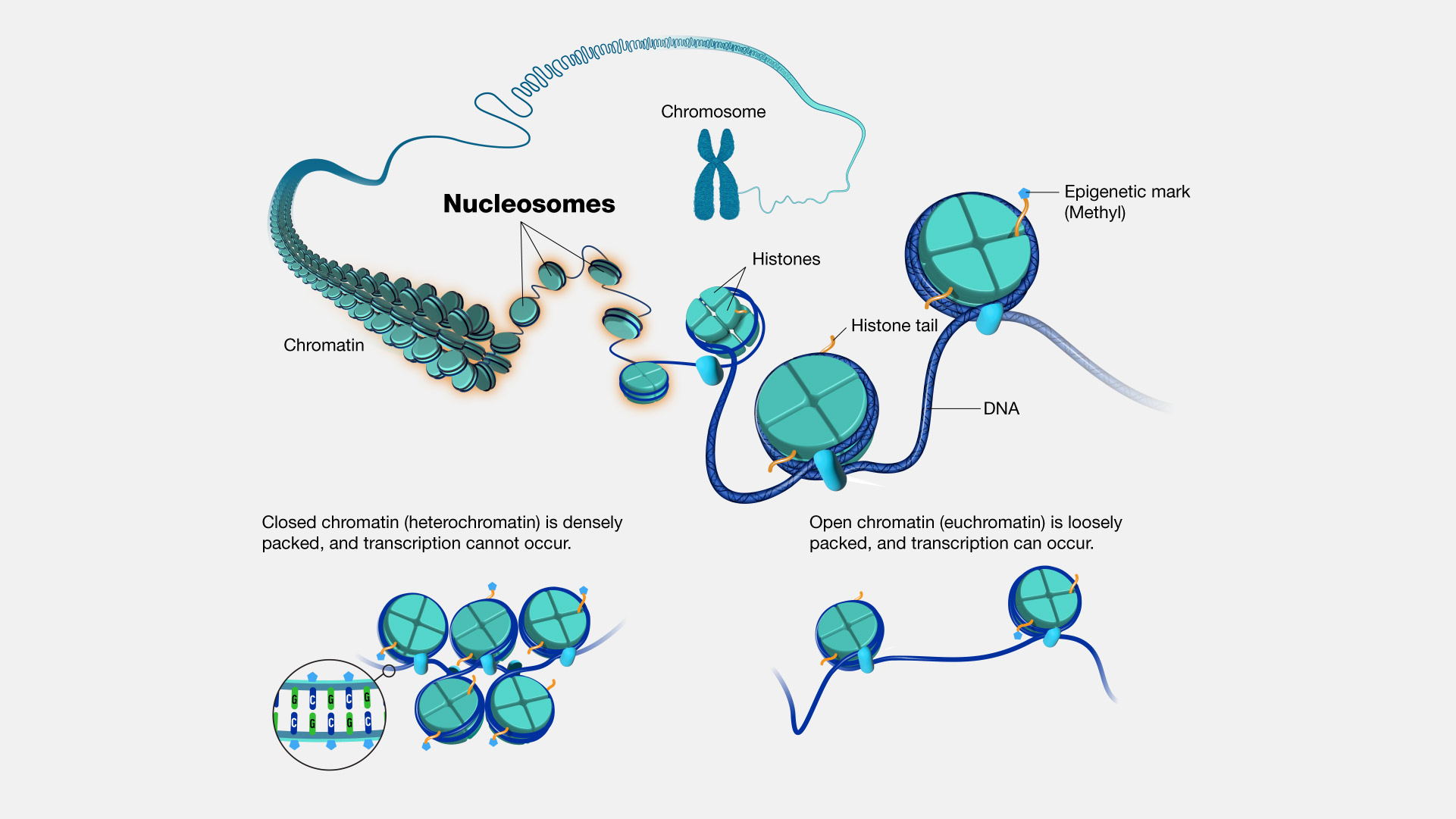
• Major remodeling complex families:
- SWI/SNF family (BAF/PBAF): Nucleosome sliding and ejection
- ISWI family: Nucleosome spacing and assembly
- CHD family: Nucleosome sliding and histone deacetylation
- INO80/SWR1 family: Histone variant exchange (H2A.Z incorporation)
• Biological roles:
- Transcriptional regulation: Controlling gene expression by modulating promoter accessibility
- DNA replication: Ensuring replication machinery access to DNA
- DNA repair: Facilitating repair protein access to damaged DNA
- Development: Orchestrating cell fate decisions and differentiation
• Clinical significance:
- Cancer: Mutations in chromatin remodelers are frequent in many cancer types
- Developmental disorders: Associated with intellectual disability and congenital abnormalities
- Aging: Dysregulation of chromatin remodeling contributes to aging phenotypes
- Therapeutic targeting: Emerging strategies for modulating chromatin remodeling in disease
• Mechanisms:
- Nucleosome sliding (moving histone proteins): Repositioning nucleosomes along DNA without disrupting histone-DNA contacts
- Histone eviction/replacement: Removing or exchanging histones to alter chromatin composition
- Histone modification: Adding or removing chemical groups that affect chromatin compaction
- ATP-dependent remodeling: Using energy from ATP hydrolysis to physically restructure chromatin

• Major remodeling complex families:
- SWI/SNF family (BAF/PBAF): Nucleosome sliding and ejection
- ISWI family: Nucleosome spacing and assembly
- CHD family: Nucleosome sliding and histone deacetylation
- INO80/SWR1 family: Histone variant exchange (H2A.Z incorporation)
• Biological roles:
- Transcriptional regulation: Controlling gene expression by modulating promoter accessibility
- DNA replication: Ensuring replication machinery access to DNA
- DNA repair: Facilitating repair protein access to damaged DNA
- Development: Orchestrating cell fate decisions and differentiation
• Clinical significance:
- Cancer: Mutations in chromatin remodelers are frequent in many cancer types
- Developmental disorders: Associated with intellectual disability and congenital abnormalities
- Aging: Dysregulation of chromatin remodeling contributes to aging phenotypes
- Therapeutic targeting: Emerging strategies for modulating chromatin remodeling in disease
🔍 Chromodomains and Bromodomains
biology
• Definition: Specialized protein that recognize specific histone modifications and mediate chromatin-based processes.
• Chromodomains:
- Structure: ~60 amino acid modules that fold into a three-stranded anti-parallel β-sheet and an α-helix
- Recognition specificity: Primarily bind to methylated lysine residues on histone tails
- Key interactions: Form an aromatic cage that accommodates the methylated lysine
- Notable examples: HP1 (binds H3K9me3), Polycomb proteins (bind H3K27me3), CHD family proteins
• Bromodomains:
- Structure: ~110 amino acid modules consisting of four α-helices forming a hydrophobic pocket
- Recognition specificity: Recognize and bind to acetylated lysine residues on histone tails
- Key interactions: Hydrogen bonding and hydrophobic interactions with the acetyl-lysine
- Notable examples: BRD family proteins, TAF1, PCAF, BRG1/BRM (in SWI/SNF complexes)
• Biological functions:
- Epigenetic regulation: Translating histone modifications into functional outcomes
- Transcriptional control: Recruiting transcriptional machinery to specific chromatin regions
- : Directing remodeling complexes to appropriate genomic locations
- DNA repair: Facilitating access of repair machinery to damaged DNA
• Applications in research and medicine:
- Epigenetic inhibitors: Bromodomain inhibitors (BETi) as emerging cancer therapeutics
- Drug discovery: Structure-based design of small molecules targeting these
- Biomarkers: Expression patterns as diagnostic or prognostic indicators
- Synthetic biology: Engineered chromatin readers for targeted gene regulation
• Chromodomains:
- Structure: ~60 amino acid modules that fold into a three-stranded anti-parallel β-sheet and an α-helix
- Recognition specificity: Primarily bind to methylated lysine residues on histone tails
- Key interactions: Form an aromatic cage that accommodates the methylated lysine
- Notable examples: HP1 (binds H3K9me3), Polycomb proteins (bind H3K27me3), CHD family proteins
• Bromodomains:
- Structure: ~110 amino acid modules consisting of four α-helices forming a hydrophobic pocket
- Recognition specificity: Recognize and bind to acetylated lysine residues on histone tails
- Key interactions: Hydrogen bonding and hydrophobic interactions with the acetyl-lysine
- Notable examples: BRD family proteins, TAF1, PCAF, BRG1/BRM (in SWI/SNF complexes)
• Biological functions:
- Epigenetic regulation: Translating histone modifications into functional outcomes
- Transcriptional control: Recruiting transcriptional machinery to specific chromatin regions
- : Directing remodeling complexes to appropriate genomic locations
- DNA repair: Facilitating access of repair machinery to damaged DNA
• Applications in research and medicine:
- Epigenetic inhibitors: Bromodomain inhibitors (BETi) as emerging cancer therapeutics
- Drug discovery: Structure-based design of small molecules targeting these
- Biomarkers: Expression patterns as diagnostic or prognostic indicators
- Synthetic biology: Engineered chromatin readers for targeted gene regulation
🧬 Cis DNA Regulatory Elements
biology
• Definition: Non-coding DNA sequences that control the transcription of nearby genes on the same chromosome by serving as binding sites for transcription factors and other regulatory proteins.
• Major types:
- Promoters: Core sequences located near transcription start sites that direct RNA polymerase binding and initiation
- Enhancers: Distal elements that increase transcription rates, often in a tissue-specific manner
- Silencers: Sequences that repress gene expression by binding negative regulatory factors
- Insulators: Boundary elements that block enhancer-promoter interactions or prevent heterochromatin spreading
- Response elements: Specific sequences that respond to environmental signals or cellular states
• Structural and functional characteristics:
- Contain specific DNA motifs recognized by transcription factors
- Can function over variable distances from target genes
- Often exhibit evolutionary conservation across species
- Frequently organized into clusters called cis-regulatory modules (CRMs)
- Can be tissue-specific, developmental stage-specific, or condition-responsive
• Identification methods:
- Comparative genomics: Identifying conserved non-coding sequences
- ChIP-seq: Mapping transcription factor binding sites genome-wide
- ATAC-seq: Identifying regions of open chromatin
- Reporter assays: Testing regulatory activity of candidate sequences
- Massively parallel reporter assays (MPRAs): High-throughput functional screening
• Biological significance:
- Orchestrate spatiotemporal gene expression patterns during development
- Mediate cellular responses to environmental stimuli
- Contribute to cell type-specific gene expression profiles
- Form the physical basis for gene regulatory networks
• Clinical and evolutionary relevance:
- Mutations in regulatory elements contribute to human disease
- Regulatory variation drives phenotypic diversity within and between species
- Therapeutic targeting of transcription factor-DNA interactions
- Synthetic biology applications in designing artificial gene circuits
• Major types:
- Promoters: Core sequences located near transcription start sites that direct RNA polymerase binding and initiation
- Enhancers: Distal elements that increase transcription rates, often in a tissue-specific manner
- Silencers: Sequences that repress gene expression by binding negative regulatory factors
- Insulators: Boundary elements that block enhancer-promoter interactions or prevent heterochromatin spreading
- Response elements: Specific sequences that respond to environmental signals or cellular states
• Structural and functional characteristics:
- Contain specific DNA motifs recognized by transcription factors
- Can function over variable distances from target genes
- Often exhibit evolutionary conservation across species
- Frequently organized into clusters called cis-regulatory modules (CRMs)
- Can be tissue-specific, developmental stage-specific, or condition-responsive
• Identification methods:
- Comparative genomics: Identifying conserved non-coding sequences
- ChIP-seq: Mapping transcription factor binding sites genome-wide
- ATAC-seq: Identifying regions of open chromatin
- Reporter assays: Testing regulatory activity of candidate sequences
- Massively parallel reporter assays (MPRAs): High-throughput functional screening
• Biological significance:
- Orchestrate spatiotemporal gene expression patterns during development
- Mediate cellular responses to environmental stimuli
- Contribute to cell type-specific gene expression profiles
- Form the physical basis for gene regulatory networks
• Clinical and evolutionary relevance:
- Mutations in regulatory elements contribute to human disease
- Regulatory variation drives phenotypic diversity within and between species
- Therapeutic targeting of transcription factor-DNA interactions
- Synthetic biology applications in designing artificial gene circuits
👁️ Convolutional Neural Network (CNN)
machine-learning
A specialized architecture inspired by the visual cortex. It uses sliding filters to automatically learn and detect important features in grid-like data (especially images), making it powerful for tasks like facial recognition, object detection, and medical image analysis.
🌐 CORS (Cross-Origin Resource Sharing)
computer scienceweb developmentnetworking
A security mechanism implemented by web browsers that allows or restricts web pages from making requests to a different , protocol, or than the one serving the web page. CORS is a relaxation of the Same-Origin Policy, which by default blocks cross-origin requests for security reasons.
How CORS Works:
When a web application running on one (e.g., https://example.com) tries to access resources from another (e.g., https://api.service.com), the browser initiates a CORS check. For simple requests, the browser adds an Origin header and checks the response for appropriate CORS headers. For complex requests, the browser first sends a preflight request (OPTIONS method) to determine if the actual request is allowed.
Key CORS Headers:
- Access-Control-Allow-Origin: Specifies which origins can access the resource
- Access-Control-Allow-Methods: Lists allowed HTTP methods (GET, POST, PUT, etc.)
- Access-Control-Allow-Headers: Specifies allowed request headers
- Access-Control-Allow-Credentials: Indicates if credentials can be included
- Access-Control-Max-Age: Sets how long preflight responses can be cached
Common CORS Scenarios:
1. API Requests: Frontend applications calling REST APIs on different
2. CDN Resources: Loading fonts, images, or scripts from content delivery networks
3. Microservices: Services communicating across different
4. Third-party Integrations: widgets or accessing external services
CORS vs Same-Origin Policy:
The Same-Origin Policy is a fundamental security concept that restricts how documents or scripts from one origin can interact with resources from another origin. CORS provides a controlled way to relax this restriction, allowing servers to specify which cross-origin requests are permitted while maintaining security.
Security Considerations:
While CORS enables legitimate cross-origin requests, misconfiguration can create security vulnerabilities. Using wildcards (*) for Access-Control-Allow-Origin with credentials, or overly permissive CORS policies can expose applications to attacks. Proper CORS configuration should follow the principle of least privilege, only allowing necessary origins and methods.
How CORS Works:
When a web application running on one (e.g., https://example.com) tries to access resources from another (e.g., https://api.service.com), the browser initiates a CORS check. For simple requests, the browser adds an Origin header and checks the response for appropriate CORS headers. For complex requests, the browser first sends a preflight request (OPTIONS method) to determine if the actual request is allowed.
Key CORS Headers:
- Access-Control-Allow-Origin: Specifies which origins can access the resource
- Access-Control-Allow-Methods: Lists allowed HTTP methods (GET, POST, PUT, etc.)
- Access-Control-Allow-Headers: Specifies allowed request headers
- Access-Control-Allow-Credentials: Indicates if credentials can be included
- Access-Control-Max-Age: Sets how long preflight responses can be cached
Common CORS Scenarios:
1. API Requests: Frontend applications calling REST APIs on different
2. CDN Resources: Loading fonts, images, or scripts from content delivery networks
3. Microservices: Services communicating across different
4. Third-party Integrations: widgets or accessing external services
CORS vs Same-Origin Policy:
The Same-Origin Policy is a fundamental security concept that restricts how documents or scripts from one origin can interact with resources from another origin. CORS provides a controlled way to relax this restriction, allowing servers to specify which cross-origin requests are permitted while maintaining security.
Security Considerations:
While CORS enables legitimate cross-origin requests, misconfiguration can create security vulnerabilities. Using wildcards (*) for Access-Control-Allow-Origin with credentials, or overly permissive CORS policies can expose applications to attacks. Proper CORS configuration should follow the principle of least privilege, only allowing necessary origins and methods.
📊 Cross Entropy Loss
machine-learningmath
A commonly used in classification problems, particularly for multi-class classification and . Cross entropy loss measures the difference between the predicted probability distribution and the true distribution (one-hot encoded labels). It penalizes confident wrong predictions more heavily than uncertain predictions.
Mathematically, for a single sample with true class y and predicted probabilities p, the cross entropy loss is:
where C is the number of classes. For binary classification, this simplifies to:
Cross entropy loss is particularly effective because it provides strong gradients when predictions are wrong and approaches zero as predictions become more accurate. It's widely used in for training on classification tasks, often combined with activation in the output layer.
Mathematically, for a single sample with true class y and predicted probabilities p, the cross entropy loss is:
where C is the number of classes. For binary classification, this simplifies to:
Cross entropy loss is particularly effective because it provides strong gradients when predictions are wrong and approaches zero as predictions become more accurate. It's widely used in for training on classification tasks, often combined with activation in the output layer.
🧪 Cytotoxicity
biology
• Definition: The degree to which a substance or agent can damage or kill living cells, typically through disruption of cellular structures, interference with metabolic pathways, or triggering of cell death mechanisms.
• Mechanisms of cytotoxicity:
- Necrosis: Uncontrolled cell death characterized by cell swelling, membrane rupture, and inflammatory response
- Apoptosis: Programmed cell death involving cell shrinkage, chromatin condensation, and formation of apoptotic bodies
- Autophagy: Self-degradation process that can lead to cell death when excessive
- Mitochondrial dysfunction: Disruption of energy production and release of pro-apoptotic factors
- DNA damage: Direct or indirect damage to genetic material leading to cell cycle arrest or death
• Detection methods:
- Membrane integrity assays: LDH release, trypan blue exclusion, propidium iodide staining
- Metabolic activity assays: MTT, MTS, XTT, WST-1, resazurin reduction
- ATP content assays: Luminescence-based detection of cellular ATP levels
- Apoptosis assays: Annexin V binding, caspase activation, TUNEL assay
- High-content imaging: Automated microscopy with multiple fluorescent markers
- Flow cytometry: Multiparametric analysis of cell death markers
• Applications in bioinformatics:
- Toxicogenomics: Computational analysis of gene expression changes in response to toxic compounds
- QSAR modeling: Predicting cytotoxicity based on chemical structure
- Pathway analysis: Identifying molecular mechanisms of toxicity
- approaches: Developing predictive models for cytotoxicity screening
- Systems biology: Integrating multi-omics data to understand cellular responses to toxicants
• Clinical and research significance:
- Drug development: Screening compounds for safety and efficacy
- Cancer research: Evaluating potential anticancer agents
- Immunology: Assessing immune cell-mediated cytotoxicity
- Environmental toxicology: Evaluating potential hazards of chemicals
- Nanomaterial safety assessment: Determining biocompatibility of engineered materials
• Mechanisms of cytotoxicity:
- Necrosis: Uncontrolled cell death characterized by cell swelling, membrane rupture, and inflammatory response
- Apoptosis: Programmed cell death involving cell shrinkage, chromatin condensation, and formation of apoptotic bodies
- Autophagy: Self-degradation process that can lead to cell death when excessive
- Mitochondrial dysfunction: Disruption of energy production and release of pro-apoptotic factors
- DNA damage: Direct or indirect damage to genetic material leading to cell cycle arrest or death
• Detection methods:
- Membrane integrity assays: LDH release, trypan blue exclusion, propidium iodide staining
- Metabolic activity assays: MTT, MTS, XTT, WST-1, resazurin reduction
- ATP content assays: Luminescence-based detection of cellular ATP levels
- Apoptosis assays: Annexin V binding, caspase activation, TUNEL assay
- High-content imaging: Automated microscopy with multiple fluorescent markers
- Flow cytometry: Multiparametric analysis of cell death markers
• Applications in bioinformatics:
- Toxicogenomics: Computational analysis of gene expression changes in response to toxic compounds
- QSAR modeling: Predicting cytotoxicity based on chemical structure
- Pathway analysis: Identifying molecular mechanisms of toxicity
- approaches: Developing predictive models for cytotoxicity screening
- Systems biology: Integrating multi-omics data to understand cellular responses to toxicants
• Clinical and research significance:
- Drug development: Screening compounds for safety and efficacy
- Cancer research: Evaluating potential anticancer agents
- Immunology: Assessing immune cell-mediated cytotoxicity
- Environmental toxicology: Evaluating potential hazards of chemicals
- Nanomaterial safety assessment: Determining biocompatibility of engineered materials
⚖️ Dalton (Da)
biology
• Definition: A unit of mass used in biochemistry and molecular biology, equivalent to 1/12 the mass of a carbon-12 atom (approximately 1.66 × 10^-24 grams).
• Equivalence: Identical to the atomic mass unit (amu), but preferred in biochemical contexts.
• Applications:
- Measuring molecular weights of proteins, nucleic acids, and other biomolecules
- Expressing the size of macromolecules (e.g., a 50 kDa protein)
- Determining mass shifts in mass spectrometry
- Calculating stoichiometry in biochemical reactions
• Context in bioinformatics:
- Protein analysis: Used to estimate protein size from amino acid sequence
- Mass spectrometry: Critical unit for peptide and protein identification
- Structural biology: Important parameter in determining molecular dimensions
- Proteomics: Used in database searching algorithms for protein identification
• Historical note: Named after John Dalton, who proposed atomic theory in the early 19th century.
• Equivalence: Identical to the atomic mass unit (amu), but preferred in biochemical contexts.
• Applications:
- Measuring molecular weights of proteins, nucleic acids, and other biomolecules
- Expressing the size of macromolecules (e.g., a 50 kDa protein)
- Determining mass shifts in mass spectrometry
- Calculating stoichiometry in biochemical reactions
• Context in bioinformatics:
- Protein analysis: Used to estimate protein size from amino acid sequence
- Mass spectrometry: Critical unit for peptide and protein identification
- Structural biology: Important parameter in determining molecular dimensions
- Proteomics: Used in database searching algorithms for protein identification
• Historical note: Named after John Dalton, who proposed atomic theory in the early 19th century.
🔬 Deconvolution Analysis
machine-learningbioinformatics
• Definition: Computational methods used to separate mixed biological signals from heterogeneous samples into their constituent components.
• Applications:
- Cell type composition estimation from bulk tissue transcriptomics data
- Tumor microenvironment characterization from mixed tumor samples
- Immune cell profiling from complex tissue samples
- Epigenetic signal deconvolution from mixed cell populations
• Key methodologies:
- Reference-based deconvolution: Uses known cell type-specific signatures as reference
- Reference-free deconvolution: Identifies cell types without prior knowledge using statistical approaches
- Semi-supervised approaches: Combines reference data with
- Spatial deconvolution: Incorporates spatial information to resolve cellular heterogeneity
• Algorithms and tools:
- CIBERSORT: Estimating immune cell fractions from gene expression profiles
- CellMix: R package for linear unmixing of heterogeneous tissue samples
- MuSiC: Multi-subject single cell deconvolution
- DSA (Digital Sorting Algorithm): Marker-free deconvolution for transcriptomics
• Challenges and considerations:
- Reference dataset quality and comprehensiveness
- Assumption of linear mixing in most algorithms
- Handling of technical and biological noise
- Validation of deconvolution results with orthogonal methods
• Applications:
- Cell type composition estimation from bulk tissue transcriptomics data
- Tumor microenvironment characterization from mixed tumor samples
- Immune cell profiling from complex tissue samples
- Epigenetic signal deconvolution from mixed cell populations
• Key methodologies:
- Reference-based deconvolution: Uses known cell type-specific signatures as reference
- Reference-free deconvolution: Identifies cell types without prior knowledge using statistical approaches
- Semi-supervised approaches: Combines reference data with
- Spatial deconvolution: Incorporates spatial information to resolve cellular heterogeneity
• Algorithms and tools:
- CIBERSORT: Estimating immune cell fractions from gene expression profiles
- CellMix: R package for linear unmixing of heterogeneous tissue samples
- MuSiC: Multi-subject single cell deconvolution
- DSA (Digital Sorting Algorithm): Marker-free deconvolution for transcriptomics
• Challenges and considerations:
- Reference dataset quality and comprehensiveness
- Assumption of linear mixing in most algorithms
- Handling of technical and biological noise
- Validation of deconvolution results with orthogonal methods
🔮 Deep Learning
machine-learning
A specialized form of using multiple layers of . It's particularly powerful for complex tasks like understanding images, text, and speech. Deep learning has enabled breakthrough applications like ChatGPT and stable diffusion models.
🦕 Deno
computer science
A modern runtime for JavaScript and TypeScript built by Ryan Dahl, the original creator of Node.js. Deno was designed to address perceived shortcomings in Node.js and provides a more secure and developer-friendly environment.
Key features include:
Security by Default: Requires explicit permissions for file, network, and environment access
TypeScript Support: Built-in TypeScript support without additional configuration
Modern APIs: Uses web-standard APIs and modern JavaScript features
No Package Manager: Imports modules directly from URLs, eliminating the need for package.json
Built-in Tools: Includes formatter, linter, test runner, and bundler
Single Executable: Distributed as a single binary with no dependencies
Deno aims to provide a more secure, simple, and modern development experience while maintaining compatibility with web standards and modern JavaScript/TypeScript practices.
Key features include:
Security by Default: Requires explicit permissions for file, network, and environment access
TypeScript Support: Built-in TypeScript support without additional configuration
Modern APIs: Uses web-standard APIs and modern JavaScript features
No Package Manager: Imports modules directly from URLs, eliminating the need for package.json
Built-in Tools: Includes formatter, linter, test runner, and bundler
Single Executable: Distributed as a single binary with no dependencies
Deno aims to provide a more secure, simple, and modern development experience while maintaining compatibility with web standards and modern JavaScript/TypeScript practices.
🔐 Diffie-Hellman Key Exchange
computer sciencemath
A cryptographic protocol that allows two parties to establish a shared secret key over an insecure communication channel without requiring a prior shared secret. It relies on the mathematical principles of modular exponentiation and the computational difficulty of the discrete logarithm problem.
The protocol works by having both parties generate private keys, derive public keys using modular exponentiation, exchange these public keys, and then independently compute the same shared secret. This method is fundamental to many secure communications systems and was the first practical implementation of public key cryptography. It's widely used in secure protocols like HTTPS, SSH, IPsec, and TLS to establish encrypted communication channels.
Video explanations:
Diffie-Hellman Key Exchange - A visual explanation of how the Diffie-Hellman protocol works and why it's secure.
The Genius Math of Modern Encryption | Diffie-Hellman Key Exchange - A visual explanation of how the Diffie-Hellman protocol works and why it's secure.
The protocol works by having both parties generate private keys, derive public keys using modular exponentiation, exchange these public keys, and then independently compute the same shared secret. This method is fundamental to many secure communications systems and was the first practical implementation of public key cryptography. It's widely used in secure protocols like HTTPS, SSH, IPsec, and TLS to establish encrypted communication channels.
Video explanations:
Diffie-Hellman Key Exchange - A visual explanation of how the Diffie-Hellman protocol works and why it's secure.
The Genius Math of Modern Encryption | Diffie-Hellman Key Exchange - A visual explanation of how the Diffie-Hellman protocol works and why it's secure.
🔄 Dimers
biology
• Definition: Molecular structures formed by the combination of two identical or similar subunits (monomers) that are joined by chemical bonds.
• Types of dimers:
- Homodimers: Composed of two identical molecular subunits
- Heterodimers: Formed from two different molecular subunits
- Protein dimers: Two protein subunits bound together (e.g., transcription factor dimers)
- Nucleic acid dimers: Paired DNA or RNA structures (e.g., thymine dimers in DNA)
• Formation mechanisms:
- Covalent bonding: Strong chemical bonds between monomers
- Non-covalent interactions: Hydrogen bonds, hydrophobic interactions, van der Waals forces
- Disulfide bridges: Covalent bonds between cysteine residues in proteins
- π-stacking: Interactions between aromatic rings
• Biological significance:
- Protein function: Many proteins are only functional as dimers
- DNA damage: Thymine dimers caused by UV radiation can lead to mutations
- Enzyme regulation: Dimerization can activate or inhibit enzymatic activity
- Signal transduction: Receptor dimerization often initiates signaling cascades
• Applications in bioinformatics:
- Protein structure prediction: Identifying potential dimerization interfaces
- Drug design: Targeting protein-protein interactions at dimer interfaces
- Sequence analysis: Detecting dimerization motifs in protein sequences
- Molecular dynamics: Simulating dimer formation and stability
• Types of dimers:
- Homodimers: Composed of two identical molecular subunits
- Heterodimers: Formed from two different molecular subunits
- Protein dimers: Two protein subunits bound together (e.g., transcription factor dimers)
- Nucleic acid dimers: Paired DNA or RNA structures (e.g., thymine dimers in DNA)
• Formation mechanisms:
- Covalent bonding: Strong chemical bonds between monomers
- Non-covalent interactions: Hydrogen bonds, hydrophobic interactions, van der Waals forces
- Disulfide bridges: Covalent bonds between cysteine residues in proteins
- π-stacking: Interactions between aromatic rings
• Biological significance:
- Protein function: Many proteins are only functional as dimers
- DNA damage: Thymine dimers caused by UV radiation can lead to mutations
- Enzyme regulation: Dimerization can activate or inhibit enzymatic activity
- Signal transduction: Receptor dimerization often initiates signaling cascades
• Applications in bioinformatics:
- Protein structure prediction: Identifying potential dimerization interfaces
- Drug design: Targeting protein-protein interactions at dimer interfaces
- Sequence analysis: Detecting dimerization motifs in protein sequences
- Molecular dynamics: Simulating dimer formation and stability
🌐 DNS (Domain Name System)
networkingcomputer scienceweb development
A hierarchical and distributed naming system that translates human-readable names into IP addresses that computers use to identify and communicate with each other on the internet. DNS acts as the internet's phone book, allowing users to access websites using memorable names instead of numerical IP addresses.
How DNS Works:
When you type a name into your browser, a DNS lookup process occurs:
1. Browser Cache: First checks if the IP address is already cached
2. Operating System Cache: Checks the local system's DNS cache
3. Router Cache: Queries the local router's DNS cache
4. ISP DNS Server: Contacts your Internet Service Provider's DNS server
5. Root Name Servers: If not found, queries one of 13 root name servers worldwide
6. TLD Name Servers: Directed to Top-Level servers (.com, .org, etc.)
7. Authoritative Name Servers: Finally queries the authoritative server for the specific
DNS Record Types:
- A Record: Maps to IPv4 address
- AAAA Record: Maps to IPv6 address
- CNAME Record: Creates an alias pointing to another
- MX Record: Specifies mail exchange servers for email delivery
- TXT Record: Stores text information (often for verification)
- NS Record: Identifies authoritative name servers for a
- PTR Record: Reverse DNS lookup (IP to )
- SOA Record: Start of Authority, contains administrative information
DNS Security:
DNS was originally designed without security in mind, leading to vulnerabilities like DNS spoofing and cache poisoning. Modern security extensions include:
- DNSSEC: Cryptographic signatures to verify DNS responses
- DNS over HTTPS (DoH): Encrypts DNS queries using HTTPS
- DNS over TLS (DoT): Encrypts DNS queries using TLS
Performance and Reliability:
DNS uses caching at multiple levels to improve performance and reduce load on authoritative servers. TTL (Time To Live) values determine how long DNS records can be cached. The distributed nature of DNS provides redundancy and fault tolerance, making it one of the most resilient systems on the internet.
How DNS Works:
When you type a name into your browser, a DNS lookup process occurs:
1. Browser Cache: First checks if the IP address is already cached
2. Operating System Cache: Checks the local system's DNS cache
3. Router Cache: Queries the local router's DNS cache
4. ISP DNS Server: Contacts your Internet Service Provider's DNS server
5. Root Name Servers: If not found, queries one of 13 root name servers worldwide
6. TLD Name Servers: Directed to Top-Level servers (.com, .org, etc.)
7. Authoritative Name Servers: Finally queries the authoritative server for the specific
DNS Record Types:
- A Record: Maps to IPv4 address
- AAAA Record: Maps to IPv6 address
- CNAME Record: Creates an alias pointing to another
- MX Record: Specifies mail exchange servers for email delivery
- TXT Record: Stores text information (often for verification)
- NS Record: Identifies authoritative name servers for a
- PTR Record: Reverse DNS lookup (IP to )
- SOA Record: Start of Authority, contains administrative information
DNS Security:
DNS was originally designed without security in mind, leading to vulnerabilities like DNS spoofing and cache poisoning. Modern security extensions include:
- DNSSEC: Cryptographic signatures to verify DNS responses
- DNS over HTTPS (DoH): Encrypts DNS queries using HTTPS
- DNS over TLS (DoT): Encrypts DNS queries using TLS
Performance and Reliability:
DNS uses caching at multiple levels to improve performance and reduce load on authoritative servers. TTL (Time To Live) values determine how long DNS records can be cached. The distributed nature of DNS provides redundancy and fault tolerance, making it one of the most resilient systems on the internet.
🏠 Domain
networkingweb-development
A human-readable address that identifies a specific location on the internet, serving as an alias for an IP address. Domains make it easier for users to access websites without memorizing complex numerical IP addresses.
A domain consists of multiple parts separated by dots, read from right to left in order of hierarchy:
- Top-Level Domain (TLD): The rightmost part (.com, .org, .net, .gov, etc.)
- Second-Level Domain: The main part of the domain name (example in example.com)
- ****: Optional prefix (www in www.example.com)
Domain Structure Example:
'.second-level.top-level' → 'www.example.com'
Common TLDs:
- .com: Commercial (most common)
- .org: Organizations
- .net: Network-related
- .edu: Educational institutions
- .gov: Government
- .io: Input/Output (popular with tech companies)
Domains are managed through the Domain Name System (DNS), which translates domain names into IP addresses that computers use to locate and connect to web servers.
A domain consists of multiple parts separated by dots, read from right to left in order of hierarchy:
- Top-Level Domain (TLD): The rightmost part (.com, .org, .net, .gov, etc.)
- Second-Level Domain: The main part of the domain name (example in example.com)
- ****: Optional prefix (www in www.example.com)
Domain Structure Example:
'.second-level.top-level' → 'www.example.com'
Common TLDs:
- .com: Commercial (most common)
- .org: Organizations
- .net: Network-related
- .edu: Educational institutions
- .gov: Government
- .io: Input/Output (popular with tech companies)
Domains are managed through the Domain Name System (DNS), which translates domain names into IP addresses that computers use to locate and connect to web servers.
📚 Edify
dictionary
To instruct or enlighten someone in a way that improves their mind, character, or understanding. The term comes from the Latin "aedificare," meaning "to build," and implies constructive learning that builds up knowledge, wisdom, or moral character. In academic and intellectual contexts, edifying content is designed to be both informative and morally or intellectually uplifting, helping individuals develop better understanding or judgment.
🔀 Elastic Net
mathmachine-learning
A hybrid regression technique that combines the penalties of both Lasso and , incorporating both L1 and L2 regularization terms. This balanced approach overcomes limitations of each method alone: it can select variables like Lasso while handling groups of correlated features better, similar to Ridge. The mixing parameter allows data scientists to tune the model between pure Lasso and pure Ridge behavior.
Elastic Net is particularly valuable for complex datasets with many correlated features, such as in genomics (where groups of genes may work together), neuroimaging (where brain regions have correlated activities), and recommendation systems (where user preferences show complex patterns).
Elastic Net is particularly valuable for complex datasets with many correlated features, such as in genomics (where groups of genes may work together), neuroimaging (where brain regions have correlated activities), and recommendation systems (where user preferences show complex patterns).
🔠 Embedding
machine-learning
A technique that converts discrete data (like words or categories) into dense vectors of continuous numbers. These learned representations capture semantic relationships and similarities, enabling AI models to process categorical data effectively. It's fundamental to modern NLP and recommendation systems.
🧬 Endogenous
dictionary
Refers to something that originates or is produced from within an organism, tissue, or cell
🧬 Epochs
machine-learning
In and , an epoch refers to one complete pass through the entire training dataset during the training process. During each epoch, the model sees every training example once and updates its parameters accordingly. Multiple epochs are typically required to train a model effectively, with the number of epochs being a hyperparameter that affects model performance and training time.
📐 Euler's Number (e)
math
A fundamental mathematical constant approximately equal to 2.71828, denoted by the letter 'e' in honor of the Swiss mathematician Leonhard Euler. It is defined as the limit of (1 + 1/n)ⁿ as n approaches infinity, or equivalently as the sum of the infinite series:
Euler's number is the base of the natural logarithm and appears naturally in many areas of mathematics, particularly in calculus where it serves as the unique number such that the derivative of eˣ equals eˣ itself. This property makes it invaluable for solving differential equations and modeling exponential growth and decay processes.
Key Applications:
- Compound Interest: Continuous compounding formula A = Pe^(rt)
- Population Growth: Exponential growth models in biology and demographics
- Radioactive Decay: Half-life calculations in physics and chemistry
- Probability Theory: Normal distribution and Poisson processes
- Signal Processing: Fourier transforms and complex analysis
- ****: Activation functions (sigmoid, ) and optimization algorithms
- Economics: Present value calculations and economic modeling
The constant e is irrational and transcendental, meaning it cannot be expressed as a simple fraction or as the root of any polynomial equation with rational coefficients. Its ubiquity in natural phenomena has earned it the designation as one of the most important mathematical constants alongside π.
Euler's number is the base of the natural logarithm and appears naturally in many areas of mathematics, particularly in calculus where it serves as the unique number such that the derivative of eˣ equals eˣ itself. This property makes it invaluable for solving differential equations and modeling exponential growth and decay processes.
Key Applications:
- Compound Interest: Continuous compounding formula A = Pe^(rt)
- Population Growth: Exponential growth models in biology and demographics
- Radioactive Decay: Half-life calculations in physics and chemistry
- Probability Theory: Normal distribution and Poisson processes
- Signal Processing: Fourier transforms and complex analysis
- ****: Activation functions (sigmoid, ) and optimization algorithms
- Economics: Present value calculations and economic modeling
The constant e is irrational and transcendental, meaning it cannot be expressed as a simple fraction or as the root of any polynomial equation with rational coefficients. Its ubiquity in natural phenomena has earned it the designation as one of the most important mathematical constants alongside π.
🔢 Exponent Rules
math
A set of fundamental algebraic rules that govern operations with exponential expressions. These rules are essential for simplifying expressions, solving equations, and working with logarithms and exponential functions.
Basic Exponent Rules:
- Product Rule: $a^m \cdot a^n = a^{m+n}$
- ****: $\frac{a^m}{a^n} = a^{m-n}$ (where $a \neq 0$)
- Power Rule: $(a^m)^n = a^{mn}$
- Power of a Product: $(ab)^n = a^n b^n$
- Power of a Quotient: $\left(\frac{a}{b}\right)^n = \frac{a^n}{b^n}$ (where $b \neq 0$)
- Zero Exponent: $a^0 = 1$ (where $a \neq 0$)
- Negative Exponent: $a^{-n} = \frac{1}{a^n}$ (where $a \neq 0$)
- Fractional Exponent: $a^{\frac{m}{n}} = \sqrt[n]{a^m} = (\sqrt[n]{a})^m$
These rules form the foundation for working with exponential and logarithmic functions, compound interest calculations, scientific notation, and are extensively used in algebra, calculus, physics, chemistry, and computer science algorithms.
Basic Exponent Rules:
- Product Rule: $a^m \cdot a^n = a^{m+n}$
- ****: $\frac{a^m}{a^n} = a^{m-n}$ (where $a \neq 0$)
- Power Rule: $(a^m)^n = a^{mn}$
- Power of a Product: $(ab)^n = a^n b^n$
- Power of a Quotient: $\left(\frac{a}{b}\right)^n = \frac{a^n}{b^n}$ (where $b \neq 0$)
- Zero Exponent: $a^0 = 1$ (where $a \neq 0$)
- Negative Exponent: $a^{-n} = \frac{1}{a^n}$ (where $a \neq 0$)
- Fractional Exponent: $a^{\frac{m}{n}} = \sqrt[n]{a^m} = (\sqrt[n]{a})^m$
These rules form the foundation for working with exponential and logarithmic functions, compound interest calculations, scientific notation, and are extensively used in algebra, calculus, physics, chemistry, and computer science algorithms.
🎯 Fine-tuning
machine-learning
The process of taking a pre-trained model and adapting it to a specific task by training it on a smaller, task-specific dataset. This transfer learning approach saves computational resources and often yields better results than training from scratch.
🔢🔍 Float (Floating-Point Number)
computer sciencemath
A float, or floating-point number, is a data type used in computer programming to represent real numbers that can have a fractional part. Unlike integers, which represent whole numbers, floats can represent a wide range of values, including very small and very large numbers, as well as numbers with decimal points. Floating-point numbers are typically stored in a format defined by the IEEE 754 standard, which specifies how to represent the number using a sign bit, an exponent, and a significand (or mantissa). Common floating-point types include single-precision (usually 32-bit) and double-precision (usually 64-bit), offering different ranges and levels of precision. While versatile, floating-point arithmetic can introduce small inaccuracies due to the finite way real numbers are approximated, leading to potential rounding errors or loss of precision in calculations.
Reference: How floating point works - jan Misali
Reference: How floating point works - jan Misali
🔬📊 Floating-Point Precision
computer sciencemath
Floating-point precision refers to the number of significant digits that can be accurately represented by a floating-point data type. It determines how close the stored floating-point number can be to the true mathematical value. Precision is limited because computers store numbers in a finite number of bits. The IEEE 754 standard defines common formats like single-precision (float) and double-precision (double). Single-precision typically offers about 7 decimal digits of precision, while double-precision offers about 15-17 decimal digits.
What this means in practice is that calculations involving floating-point numbers may not always be exact. For example, representing 0.1 in binary floating-point is not perfectly accurate, similar to how 1/3 cannot be perfectly represented as a finite decimal. This can lead to:
- Rounding Errors: Small discrepancies that occur when a number is rounded to fit the available precision.
- Loss of Significance: When subtracting two nearly equal numbers, significant digits can be lost, leading to a result with much lower relative accuracy.
- Comparison Issues: Directly comparing two floating-point numbers for equality (e.g., `a == b`) can be unreliable due to these small precision differences. It's often better to check if their absolute difference is within a small tolerance (epsilon).
Understanding floating-point precision is crucial in scientific computing, financial calculations, and any where numerical accuracy is important, as ignoring these limitations can lead to incorrect results or unexpected behavior in programs.
Reference: How floating point works - jan Misali
What this means in practice is that calculations involving floating-point numbers may not always be exact. For example, representing 0.1 in binary floating-point is not perfectly accurate, similar to how 1/3 cannot be perfectly represented as a finite decimal. This can lead to:
- Rounding Errors: Small discrepancies that occur when a number is rounded to fit the available precision.
- Loss of Significance: When subtracting two nearly equal numbers, significant digits can be lost, leading to a result with much lower relative accuracy.
- Comparison Issues: Directly comparing two floating-point numbers for equality (e.g., `a == b`) can be unreliable due to these small precision differences. It's often better to check if their absolute difference is within a small tolerance (epsilon).
Understanding floating-point precision is crucial in scientific computing, financial calculations, and any where numerical accuracy is important, as ignoring these limitations can lead to incorrect results or unexpected behavior in programs.
Reference: How floating point works - jan Misali
🧬 Gene Fusion Events
biology
• Definition: Hybrid genes formed by the joining of two previously separate genes, typically resulting from chromosomal rearrangements such as translocations, inversions, or deletions.
• Formation mechanisms:
- Chromosomal translocations: Exchange of genetic material between non-homologous chromosomes
- Chromosomal inversions: Reversal of a DNA segment within a chromosome
- Tandem duplications: Duplication of a segment followed by fusion
- Transcription-mediated gene fusion: Read-through transcription between adjacent genes
- Trans-splicing: Joining of exons from separate pre-mRNA molecules
• Structural characteristics:
- Breakpoint junctions: Points where the two genes are joined
- Fusion : Protein contributed by each partner gene
- Reading frame: Determines if the fusion produces a functional protein
- Regulatory elements: Promoters and enhancers that control fusion gene expression
• Detection methods:
- RNA-seq with fusion-detection algorithms (STAR-Fusion, FusionCatcher)
- Whole genome sequencing to identify genomic breakpoints
- FISH (Fluorescence In Situ Hybridization) for known fusions
- RT-PCR with fusion-specific primers
- Mass spectrometry for fusion protein detection
• Biological and clinical significance:
- Oncogenic drivers in many cancer types (e.g., BCR-ABL in chronic myeloid leukemia)
- Diagnostic biomarkers for cancer classification
- Therapeutic targets for precision medicine approaches
- Evolutionary mechanism for new gene function
- Contribution to genetic diversity and adaptation
• Notable examples:
- BCR-ABL1 in chronic myeloid leukemia (Phil Philadelphia chromosome)
- EML4-ALK in non-small cell lung cancer
- TMPRSS2-ERG in prostate cancer
- PML-RARA in acute promyelocytic leukemia
- SYT-SSX in synovial sarcoma
• Formation mechanisms:
- Chromosomal translocations: Exchange of genetic material between non-homologous chromosomes
- Chromosomal inversions: Reversal of a DNA segment within a chromosome
- Tandem duplications: Duplication of a segment followed by fusion
- Transcription-mediated gene fusion: Read-through transcription between adjacent genes
- Trans-splicing: Joining of exons from separate pre-mRNA molecules
• Structural characteristics:
- Breakpoint junctions: Points where the two genes are joined
- Fusion : Protein contributed by each partner gene
- Reading frame: Determines if the fusion produces a functional protein
- Regulatory elements: Promoters and enhancers that control fusion gene expression
• Detection methods:
- RNA-seq with fusion-detection algorithms (STAR-Fusion, FusionCatcher)
- Whole genome sequencing to identify genomic breakpoints
- FISH (Fluorescence In Situ Hybridization) for known fusions
- RT-PCR with fusion-specific primers
- Mass spectrometry for fusion protein detection
• Biological and clinical significance:
- Oncogenic drivers in many cancer types (e.g., BCR-ABL in chronic myeloid leukemia)
- Diagnostic biomarkers for cancer classification
- Therapeutic targets for precision medicine approaches
- Evolutionary mechanism for new gene function
- Contribution to genetic diversity and adaptation
• Notable examples:
- BCR-ABL1 in chronic myeloid leukemia (Phil Philadelphia chromosome)
- EML4-ALK in non-small cell lung cancer
- TMPRSS2-ERG in prostate cancer
- PML-RARA in acute promyelocytic leukemia
- SYT-SSX in synovial sarcoma
🌐 Generalized CP (GCP) Decomposition
mathmachine-learning
An extension of the standard CP decomposition that incorporates different and constraints to handle various data types (binary, count, continuous) and missing values. GCP provides more flexibility for modeling complex real-world data with non-Gaussian characteristics.
In , GCP enables robust pattern discovery in heterogeneous multi-way data, supporting applications like topic modeling across document collections, community detection in dynamic networks, and analyzing sparse, noisy biological measurements across multiple experimental conditions.
In , GCP enables robust pattern discovery in heterogeneous multi-way data, supporting applications like topic modeling across document collections, community detection in dynamic networks, and analyzing sparse, noisy biological measurements across multiple experimental conditions.
🎨 Generative Adversarial Network (GAN)
machine-learning
An AI architecture where two networks compete: one creates fake data, while the other tries to distinguish real from fake. This competition drives both to improve, resulting in increasingly realistic synthetic data. GANs have revolutionized AI-generated art, deepfakes, and synthetic data generation.
🔍✅ Git Triage:
git
The process of reviewing, labeling, categorizing, and prioritizing issues and pull requests in a Git repository to ensure effective project management and workflow. Git triage helps maintain order, identify critical tasks, eliminate duplicates, and streamline collaboration among contributors.
⬇️ Gradient Descent
machine-learningmath
A fundamental optimization algorithm used to train models by iteratively adjusting parameters to minimize a . The algorithm computes the gradient (partial derivatives) of the with respect to each parameter and updates parameters in the direction opposite to the gradient, effectively moving downhill toward a minimum.
Mathematically, the update rule is:
where θ represents parameters, α is the learning rate, and ∇J(θ) is the gradient of the .
Main Variants:
Batch Gradient Descent: Uses the entire dataset to compute gradients at each step. Provides stable convergence but can be computationally expensive for large datasets.
** Gradient Descent (SGD)**: Uses a single random sample to compute gradients at each step. Much faster per iteration and can escape local minima due to noise, but convergence is more erratic.
Mini-batch Gradient Descent: Uses small batches of samples (typically 32-256) to compute gradients. Balances the stability of batch gradient descent with the efficiency of SGD, making it the most commonly used variant in practice.
Gradient descent is the foundation of most optimization and is essential for training , linear regression, logistic regression, and many other models.
Mathematically, the update rule is:
where θ represents parameters, α is the learning rate, and ∇J(θ) is the gradient of the .
Main Variants:
Batch Gradient Descent: Uses the entire dataset to compute gradients at each step. Provides stable convergence but can be computationally expensive for large datasets.
** Gradient Descent (SGD)**: Uses a single random sample to compute gradients at each step. Much faster per iteration and can escape local minima due to noise, but convergence is more erratic.
Mini-batch Gradient Descent: Uses small batches of samples (typically 32-256) to compute gradients. Balances the stability of batch gradient descent with the efficiency of SGD, making it the most commonly used variant in practice.
Gradient descent is the foundation of most optimization and is essential for training , linear regression, logistic regression, and many other models.
🧠 Hebbian Theory
biology
• Definition: A neurobiological theory that explains how synaptic connections between neurons are strengthened through repeated and persistent stimulation, often summarized as "cells that fire together, wire together."
• Core principle:
- Synaptic plasticity: When two neurons are repeatedly active at the same time, the synaptic connection between them becomes stronger
- Activity-dependent modification: The strength of neural connections depends on the correlation of activity between pre- and post-synaptic neurons
- Learning mechanism: Forms the basis for associative learning and memory formation
• Mathematical formulation:
- Hebbian learning rule: Δw = η × x × y (where Δw is the change in synaptic weight, η is the learning rate, x is pre-synaptic activity, y is post-synaptic activity)
- Long-term potentiation (LTP): Persistent strengthening of synapses based on recent patterns of activity
- Long-term depression (LTD): Weakening of synaptic connections when neurons fire out of sync
• Applications in computational neuroscience:
- Artificial : Hebbian learning algorithms for
- Self-organizing maps: architectures that use Hebbian principles for pattern recognition
- Spike-timing dependent plasticity (STDP): Refined models that consider the precise timing of neural spikes
- Memory models: Computational frameworks for understanding how memories are stored and retrieved
• Biological significance:
- Development: Critical for proper neural circuit formation during brain development
- Learning and memory: Fundamental mechanism underlying associative learning and memory consolidation
- Adaptation: Allows neural circuits to adapt to environmental changes and experiences
- Pathology: Dysregulation implicated in neurodevelopmental disorders and neurodegenerative diseases
• Modern extensions:
- Spike-timing dependent plasticity: Considers the precise timing of pre- and post-synaptic spikes
- Homeostatic plasticity: Mechanisms that maintain overall neural activity within functional ranges
- Metaplasticity: Changes in the ability to induce synaptic plasticity based on prior synaptic activity
- Computational implementations: algorithms inspired by Hebbian principles
• Core principle:
- Synaptic plasticity: When two neurons are repeatedly active at the same time, the synaptic connection between them becomes stronger
- Activity-dependent modification: The strength of neural connections depends on the correlation of activity between pre- and post-synaptic neurons
- Learning mechanism: Forms the basis for associative learning and memory formation
• Mathematical formulation:
- Hebbian learning rule: Δw = η × x × y (where Δw is the change in synaptic weight, η is the learning rate, x is pre-synaptic activity, y is post-synaptic activity)
- Long-term potentiation (LTP): Persistent strengthening of synapses based on recent patterns of activity
- Long-term depression (LTD): Weakening of synaptic connections when neurons fire out of sync
• Applications in computational neuroscience:
- Artificial : Hebbian learning algorithms for
- Self-organizing maps: architectures that use Hebbian principles for pattern recognition
- Spike-timing dependent plasticity (STDP): Refined models that consider the precise timing of neural spikes
- Memory models: Computational frameworks for understanding how memories are stored and retrieved
• Biological significance:
- Development: Critical for proper neural circuit formation during brain development
- Learning and memory: Fundamental mechanism underlying associative learning and memory consolidation
- Adaptation: Allows neural circuits to adapt to environmental changes and experiences
- Pathology: Dysregulation implicated in neurodevelopmental disorders and neurodegenerative diseases
• Modern extensions:
- Spike-timing dependent plasticity: Considers the precise timing of pre- and post-synaptic spikes
- Homeostatic plasticity: Mechanisms that maintain overall neural activity within functional ranges
- Metaplasticity: Changes in the ability to induce synaptic plasticity based on prior synaptic activity
- Computational implementations: algorithms inspired by Hebbian principles
🧩 Hi-C (Chromosome Conformation Capture + Sequencing)
biologylab-techniques
• Purpose: Reveals 3D organization of the genome by capturing DNA-DNA interactions across long distances.
• Data type: Contact maps showing spatial proximity between regions of the genome.
• Used for: Studying chromatin loops, compartments, and topologically associating (TADs).
| Method | Focus | Insight Gained |
|------------|--------------------------------|-------------------------------------------------|
| RNA-seq | Transcribed RNA | Which genes are active and how much |
| ATAC-seq | Open chromatin | Where DNA is accessible and likely regulatory |
| ChIP-seq | Protein-DNA binding | Where transcription factors or histones bind |
| Hi-C | 3D genome architecture | How DNA is folded and organized in the nucleus |
• Data type: Contact maps showing spatial proximity between regions of the genome.
• Used for: Studying chromatin loops, compartments, and topologically associating (TADs).
| Method | Focus | Insight Gained |
|------------|--------------------------------|-------------------------------------------------|
| RNA-seq | Transcribed RNA | Which genes are active and how much |
| ATAC-seq | Open chromatin | Where DNA is accessible and likely regulatory |
| ChIP-seq | Protein-DNA binding | Where transcription factors or histones bind |
| Hi-C | 3D genome architecture | How DNA is folded and organized in the nucleus |
🧬 HLA Imputation
biology
• Definition: Computational methods to predict human leukocyte antigen (HLA) genotypes from SNP data or low-coverage sequencing, leveraging linkage disequilibrium patterns.
• Importance of HLA:
- Critical role in immune response and self/non-self recognition
- Highly polymorphic gene family with thousands of alleles
- Strong association with autoimmune diseases, transplant outcomes, and drug hypersensitivity
- Relevant for vaccine development and immunotherapy response prediction
• Imputation approaches:
- Statistical methods: Using reference panels and linkage disequilibrium patterns
- : and other ML approaches for HLA type prediction
- Graph-based methods: Leveraging haplotype structures and population-specific patterns
- Hybrid approaches: Combining multiple methods for improved accuracy
• Tools and resources:
- SNP2HLA: Imputes classical HLA alleles and amino acid polymorphisms
- HIBAG: HLA genotype imputation with attribute bagging
- HLA*IMP: Statistical HLA imputation using reference panels
- T1DGC and 1000 Genomes reference panels: Population-specific HLA haplotype references
• Applications:
- GWAS: Fine-mapping of disease associations in the HLA region
- Pharmacogenomics: Predicting adverse drug reactions
- Transplantation medicine: Virtual crossmatching and donor selection
- Population genetics: Studying HLA diversity across human populations
• Importance of HLA:
- Critical role in immune response and self/non-self recognition
- Highly polymorphic gene family with thousands of alleles
- Strong association with autoimmune diseases, transplant outcomes, and drug hypersensitivity
- Relevant for vaccine development and immunotherapy response prediction
• Imputation approaches:
- Statistical methods: Using reference panels and linkage disequilibrium patterns
- : and other ML approaches for HLA type prediction
- Graph-based methods: Leveraging haplotype structures and population-specific patterns
- Hybrid approaches: Combining multiple methods for improved accuracy
• Tools and resources:
- SNP2HLA: Imputes classical HLA alleles and amino acid polymorphisms
- HIBAG: HLA genotype imputation with attribute bagging
- HLA*IMP: Statistical HLA imputation using reference panels
- T1DGC and 1000 Genomes reference panels: Population-specific HLA haplotype references
• Applications:
- GWAS: Fine-mapping of disease associations in the HLA region
- Pharmacogenomics: Predicting adverse drug reactions
- Transplantation medicine: Virtual crossmatching and donor selection
- Population genetics: Studying HLA diversity across human populations
🧬 Homologs
biology
• Definition: Genes that share a common evolutionary ancestry, regardless of whether they arose through speciation () or duplication ().
• Types of homology relationships:
- : Genes separated by speciation
- : Genes separated by duplication
- : Genes acquired through horizontal gene transfer
- Ohnologs: resulting from whole genome duplication
• Detection methods:
- Sequence similarity searches (BLAST, HMMER)
- Protein architecture analysis
- 3D structural comparison
- Combined phylogenetic and genomic approaches
• Degrees of homology:
- Complete homology: Similarity across entire sequence length
- Partial homology: Similarity in specific or regions
- Remote homology: Detectable only through sensitive methods or structural analysis
• Applications in research:
- Inferring protein function and structure
- Reconstructing gene and genome evolution
- Understanding protein family expansion and contraction
- Identifying conserved functional sites
• Types of homology relationships:
- : Genes separated by speciation
- : Genes separated by duplication
- : Genes acquired through horizontal gene transfer
- Ohnologs: resulting from whole genome duplication
• Detection methods:
- Sequence similarity searches (BLAST, HMMER)
- Protein architecture analysis
- 3D structural comparison
- Combined phylogenetic and genomic approaches
• Degrees of homology:
- Complete homology: Similarity across entire sequence length
- Partial homology: Similarity in specific or regions
- Remote homology: Detectable only through sensitive methods or structural analysis
• Applications in research:
- Inferring protein function and structure
- Reconstructing gene and genome evolution
- Understanding protein family expansion and contraction
- Identifying conserved functional sites
🌐 Internet Protocol (IP)
networkingcomputer science
A fundamental communication protocol that governs how data is transmitted across networks, including the internet. The Internet Protocol is part of the Internet Protocol Suite (TCP/IP) and operates at the network layer (Layer 3) of the OSI model.
IP is responsible for addressing, routing, and delivering data packets between devices across interconnected networks. It provides a standardized method for devices to communicate regardless of their underlying hardware or network technologies.
Key Functions:
- Addressing: Assigns unique IP addresses to identify devices on networks
- Routing: Determines the best path for data packets to travel from source to destination
- Packet Forwarding: Moves data packets through intermediate routers and networks
- Fragmentation and Reassembly: Breaks large data into smaller packets and reconstructs them at the destination
IP Versions:
IPv4 (Internet Protocol version 4):
- Address Format: 32-bit addresses (e.g., 192.168.1.1)
- Address Space: ~4.3 billion unique addresses
- Status: Widely deployed but facing address exhaustion
- Header Size: 20-60 bytes
IPv6 (Internet Protocol version 6):
- Address Format: 128-bit addresses (e.g., 2001:db8::1)
- Address Space: ~340 undecillion addresses
- Status: Designed to replace IPv4 and solve scalability issues
- Header Size: 40 bytes (fixed)
- Features: Built-in security, better QoS support, simplified header structure
IP Characteristics:
- Connectionless: Each packet is treated independently
- Best Effort: No guarantee of delivery, ordering, or error correction
- Unreliable: Higher-layer protocols (like TCP) provide reliability when needed
IP forms the foundation of internet communication, enabling global connectivity and the modern digital world.
IP is responsible for addressing, routing, and delivering data packets between devices across interconnected networks. It provides a standardized method for devices to communicate regardless of their underlying hardware or network technologies.
Key Functions:
- Addressing: Assigns unique IP addresses to identify devices on networks
- Routing: Determines the best path for data packets to travel from source to destination
- Packet Forwarding: Moves data packets through intermediate routers and networks
- Fragmentation and Reassembly: Breaks large data into smaller packets and reconstructs them at the destination
IP Versions:
IPv4 (Internet Protocol version 4):
- Address Format: 32-bit addresses (e.g., 192.168.1.1)
- Address Space: ~4.3 billion unique addresses
- Status: Widely deployed but facing address exhaustion
- Header Size: 20-60 bytes
IPv6 (Internet Protocol version 6):
- Address Format: 128-bit addresses (e.g., 2001:db8::1)
- Address Space: ~340 undecillion addresses
- Status: Designed to replace IPv4 and solve scalability issues
- Header Size: 40 bytes (fixed)
- Features: Built-in security, better QoS support, simplified header structure
IP Characteristics:
- Connectionless: Each packet is treated independently
- Best Effort: No guarantee of delivery, ordering, or error correction
- Unreliable: Higher-layer protocols (like TCP) provide reliability when needed
IP forms the foundation of internet communication, enabling global connectivity and the modern digital world.
🌐 IP Address (Internet Protocol Address)
networking
A unique numerical identifier assigned to every device connected to a computer network that uses the Internet Protocol for communication. IP addresses serve as the fundamental addressing system that enables devices to locate and communicate with each other across networks.
IP addresses function like postal addresses for the internet, allowing data packets to be routed from source to destination. They are essential for all internet communication, from web browsing to email to file transfers.
IPv4 (Internet Protocol version 4):
- Format: Four decimal numbers (0-255) separated by dots
- Example: 192.168.1.1 or 8.8.8.8
- Address Space: ~4.3 billion unique addresses (2³²)
- Structure: 32-bit addresses written in dotted decimal notation
- Status: Widely used but address space is nearly exhausted
IPv6 (Internet Protocol version 6):
- Format: Eight groups of four hexadecimal digits separated by colons
- Example: 2001:0db8:85a3:0000:0000:8a2e:0370:7334
- Address Space: ~340 undecillion addresses (2¹²⁸)
- Structure: 128-bit addresses with shorthand notation support
- Status: Designed to replace IPv4 and solve address exhaustion
Types of IP Addresses:
- Public: Globally unique, routable on the internet
- Private: Used within local networks (192.168.x.x, 10.x.x.x, 172.16-31.x.x)
- Static: Permanently assigned to a device
- Dynamic: Temporarily assigned by DHCP servers
Key Functions:
- Device identification and location
- Network routing and packet delivery
- Network security and access control
- Geolocation and content delivery optimization
IP addresses function like postal addresses for the internet, allowing data packets to be routed from source to destination. They are essential for all internet communication, from web browsing to email to file transfers.
IPv4 (Internet Protocol version 4):
- Format: Four decimal numbers (0-255) separated by dots
- Example: 192.168.1.1 or 8.8.8.8
- Address Space: ~4.3 billion unique addresses (2³²)
- Structure: 32-bit addresses written in dotted decimal notation
- Status: Widely used but address space is nearly exhausted
IPv6 (Internet Protocol version 6):
- Format: Eight groups of four hexadecimal digits separated by colons
- Example: 2001:0db8:85a3:0000:0000:8a2e:0370:7334
- Address Space: ~340 undecillion addresses (2¹²⁸)
- Structure: 128-bit addresses with shorthand notation support
- Status: Designed to replace IPv4 and solve address exhaustion
Types of IP Addresses:
- Public: Globally unique, routable on the internet
- Private: Used within local networks (192.168.x.x, 10.x.x.x, 172.16-31.x.x)
- Static: Permanently assigned to a device
- Dynamic: Temporarily assigned by DHCP servers
Key Functions:
- Device identification and location
- Network routing and packet delivery
- Network security and access control
- Geolocation and content delivery optimization
🧬 Isoforms
biology
• Definition: Different forms of a protein that arise from the same gene through , alternative promoter usage, or alternative polyadenylation.
• Generation mechanisms:
- : Inclusion or exclusion of specific exons
- Alternative promoter usage: Transcription initiation from different start sites
- Alternative polyadenylation: Different 3' end processing of mRNA
- RNA editing: Post-transcriptional modification of nucleotides
• Biological significance:
- Increases proteome diversity without expanding the genome
- Enables tissue-specific or developmental stage-specific protein functions
- Allows of protein activity, localization, or interaction partners
- Contributes to phenotypic complexity in higher organisms
• Characteristics:
- Share some sequence regions but differ in others
- May have different functional , subcellular localization signals, or regulatory sites
- Often exhibit tissue-specific or condition-specific expression patterns
- Can have distinct or overlapping functions
• Detection methods:
- RNA-seq with isoform-specific analysis
- Long-read sequencing (PacBio, Nanopore)
- Isoform-specific RT-PCR
- Mass spectrometry-based proteomics
• Clinical relevance:
- Aberrant isoform expression associated with various diseases
- Cancer-specific isoforms as potential biomarkers or therapeutic targets
- Genetic variants affecting splicing linked to hereditary disorders
- Isoform-specific drug targeting strategies
• Generation mechanisms:
- : Inclusion or exclusion of specific exons
- Alternative promoter usage: Transcription initiation from different start sites
- Alternative polyadenylation: Different 3' end processing of mRNA
- RNA editing: Post-transcriptional modification of nucleotides
• Biological significance:
- Increases proteome diversity without expanding the genome
- Enables tissue-specific or developmental stage-specific protein functions
- Allows of protein activity, localization, or interaction partners
- Contributes to phenotypic complexity in higher organisms
• Characteristics:
- Share some sequence regions but differ in others
- May have different functional , subcellular localization signals, or regulatory sites
- Often exhibit tissue-specific or condition-specific expression patterns
- Can have distinct or overlapping functions
• Detection methods:
- RNA-seq with isoform-specific analysis
- Long-read sequencing (PacBio, Nanopore)
- Isoform-specific RT-PCR
- Mass spectrometry-based proteomics
• Clinical relevance:
- Aberrant isoform expression associated with various diseases
- Cancer-specific isoforms as potential biomarkers or therapeutic targets
- Genetic variants affecting splicing linked to hereditary disorders
- Isoform-specific drug targeting strategies
🧪 Kinases
biology
• Definition: Enzymes that catalyze the transfer of phosphate groups from high-energy, phosphate-donating molecules (typically ATP) to specific substrates, including proteins, lipids, and nucleic acids, in a process known as phosphorylation.
• Types and classification:
- Protein kinases: Phosphorylate proteins on serine, threonine, or tyrosine residues
- Lipid kinases: Add phosphate groups to lipids (e.g., phosphoinositide kinases)
- Carbohydrate kinases: Phosphorylate sugars during metabolic processes
- Nucleotide kinases: Add phosphate groups to nucleosides to form nucleotides
• Protein kinase families:
- Serine/threonine kinases: Phosphorylate the hydroxyl groups of serine or threonine residues
- Tyrosine kinases: Phosphorylate tyrosine residues (e.g., receptor tyrosine kinases like EGFR, PDGFR)
- Dual-specificity kinases: Can phosphorylate serine, threonine, and tyrosine residues
- Histidine kinases: Primarily found in prokaryotes and plants, phosphorylate histidine residues
• Biological functions:
- Signal transduction: Transmitting signals from cell surface receptors to intracellular targets
- Metabolic regulation: Controlling enzymatic activity in metabolic pathways
- Cell cycle control: Regulating progression through different phases of the cell cycle
- Gene expression: Modifying transcription factors to alter gene expression patterns
- Protein function: Altering protein activity, stability, localization, or interactions
• Mechanism of action:
- Binding of ATP and substrate within the catalytic
- Transfer of the γ-phosphate group from ATP to the target molecule
- Release of the phosphorylated substrate and ADP
- Regulation through various mechanisms including phosphorylation, protein interactions, and conformational changes
• Structural features:
- Conserved catalytic with glycine-rich ATP-binding region
- Activation loop that regulates kinase activity
- Substrate-binding regions that determine specificity
- Regulatory that control kinase function
• Clinical significance:
- Dysregulation implicated in numerous diseases including cancer, inflammatory disorders, and neurological conditions
- Important drug targets with over 50 FDA-approved kinase inhibitors
- Examples include imatinib (targets Abelson tyrosine kinase in chronic myelogenous leukemia)
- Emerging approaches include targeted degradation of kinases using PROTACs or molecular glue degraders
• Bioinformatic applications:
- Kinome analysis: Computational study of the complete set of kinases in an organism
- Prediction of phosphorylation sites using algorithms
- Structural modeling to design selective kinase inhibitors
- Network analysis of kinase signaling pathways in disease states
• Types and classification:
- Protein kinases: Phosphorylate proteins on serine, threonine, or tyrosine residues
- Lipid kinases: Add phosphate groups to lipids (e.g., phosphoinositide kinases)
- Carbohydrate kinases: Phosphorylate sugars during metabolic processes
- Nucleotide kinases: Add phosphate groups to nucleosides to form nucleotides
• Protein kinase families:
- Serine/threonine kinases: Phosphorylate the hydroxyl groups of serine or threonine residues
- Tyrosine kinases: Phosphorylate tyrosine residues (e.g., receptor tyrosine kinases like EGFR, PDGFR)
- Dual-specificity kinases: Can phosphorylate serine, threonine, and tyrosine residues
- Histidine kinases: Primarily found in prokaryotes and plants, phosphorylate histidine residues
• Biological functions:
- Signal transduction: Transmitting signals from cell surface receptors to intracellular targets
- Metabolic regulation: Controlling enzymatic activity in metabolic pathways
- Cell cycle control: Regulating progression through different phases of the cell cycle
- Gene expression: Modifying transcription factors to alter gene expression patterns
- Protein function: Altering protein activity, stability, localization, or interactions
• Mechanism of action:
- Binding of ATP and substrate within the catalytic
- Transfer of the γ-phosphate group from ATP to the target molecule
- Release of the phosphorylated substrate and ADP
- Regulation through various mechanisms including phosphorylation, protein interactions, and conformational changes
• Structural features:
- Conserved catalytic with glycine-rich ATP-binding region
- Activation loop that regulates kinase activity
- Substrate-binding regions that determine specificity
- Regulatory that control kinase function
• Clinical significance:
- Dysregulation implicated in numerous diseases including cancer, inflammatory disorders, and neurological conditions
- Important drug targets with over 50 FDA-approved kinase inhibitors
- Examples include imatinib (targets Abelson tyrosine kinase in chronic myelogenous leukemia)
- Emerging approaches include targeted degradation of kinases using PROTACs or molecular glue degraders
• Bioinformatic applications:
- Kinome analysis: Computational study of the complete set of kinases in an organism
- Prediction of phosphorylation sites using algorithms
- Structural modeling to design selective kinase inhibitors
- Network analysis of kinase signaling pathways in disease states
📊 Lasso Regression
mathmachine-learning
A linear regression technique that performs both variable selection and regularization to enhance prediction accuracy and interpretability. Lasso (Least Absolute Shrinkage and Selection Operator) adds an L1 penalty term to the cost function, which can shrink some coefficients exactly to zero, effectively removing less important features from the model. This feature selection capability makes Lasso particularly valuable for high-dimensional datasets where many features may be irrelevant or redundant.
Lasso Regression is widely used in fields like genomics (selecting relevant genetic markers), finance (identifying key economic indicators), and image processing (extracting important features while discarding noise).
Lasso Regression is widely used in fields like genomics (selecting relevant genetic markers), finance (identifying key economic indicators), and image processing (extracting important features while discarding noise).
🧠 Long Short-Term Memory (LSTM)
machine-learning
An advanced RNN architecture that solves the "vanishing gradient" problem, allowing it to remember important information for longer sequences. It uses specialized gates to control information flow, making it excellent for tasks requiring long-term memory like language translation and speech recognition.
📊 Loss Function
mathmachine-learning
A mathematical function that measures how far a model's predictions are from the actual target values, providing a quantifiable way to assess model performance during training. The loss function calculates the "cost" or "error" of the model's current state, with lower values indicating better performance. Different types of problems require different loss functions: mean squared error for regression tasks, cross-entropy for classification, and specialized losses for tasks like object detection or generative modeling.
The choice of loss function is crucial as it directly influences how the model learns through optimization. During training, the algorithm adjusts model parameters to minimize the loss function, effectively teaching the model to make better predictions. Common examples include mean absolute error (L1 loss), mean squared error (L2 loss), binary cross-entropy, categorical cross-entropy, and Huber loss. Modern often employs custom loss functions tailored to specific tasks, such as focal loss for handling class imbalance or perceptual loss for image generation tasks.
The choice of loss function is crucial as it directly influences how the model learns through optimization. During training, the algorithm adjusts model parameters to minimize the loss function, effectively teaching the model to make better predictions. Common examples include mean absolute error (L1 loss), mean squared error (L2 loss), binary cross-entropy, categorical cross-entropy, and Huber loss. Modern often employs custom loss functions tailored to specific tasks, such as focal loss for handling class imbalance or perceptual loss for image generation tasks.
🤖 Machine Learning
machine-learning
A subset of AI where systems improve their performance on a task through experience (data), without being explicitly programmed. It's crucial for applications like image recognition, recommendation systems, and natural language processing.
🔬 Mechanistic Modeling
machine-learning
An approach to modeling that incorporates known physical, chemical, or biological processes to explain phenomena, rather than relying solely on statistical patterns in data. Unlike purely data-driven approaches, mechanistic models are built on theoretical understanding of the underlying mechanisms that generate the observed data. These models attempt to represent causal relationships and system dynamics based on first principles.
Mechanistic models are particularly valuable in scientific where understanding the "why" and "how" is as important as prediction accuracy. They offer greater interpretability and can often extrapolate beyond the range of observed data more reliably than black-box statistical models. In , mechanistic approaches are increasingly being combined with data-driven methods to create hybrid models that benefit from both theoretical knowledge and empirical patterns, especially in fields like computational biology, climate science, and physics-informed .
Flight Simulator:
A flight simulator uses mathematical equations to recreate the experience of flying a plane, demonstrating a real-world application of a mechanistic model.
Chromatography Model:
In chromatography, mechanistic models consider physical and biochemical effects like convection, dispersion, and adsorption, based on natural laws
A model (e.g., a ) can be trained to predict cell phenotypes (e.g., cell growth, cell death) based on the outputs of the mechanistic model, which include changes in protein localization, phosphorylation, and downstream effects
Mechanistic models are particularly valuable in scientific where understanding the "why" and "how" is as important as prediction accuracy. They offer greater interpretability and can often extrapolate beyond the range of observed data more reliably than black-box statistical models. In , mechanistic approaches are increasingly being combined with data-driven methods to create hybrid models that benefit from both theoretical knowledge and empirical patterns, especially in fields like computational biology, climate science, and physics-informed .
Flight Simulator:
A flight simulator uses mathematical equations to recreate the experience of flying a plane, demonstrating a real-world application of a mechanistic model.
Chromatography Model:
In chromatography, mechanistic models consider physical and biochemical effects like convection, dispersion, and adsorption, based on natural laws
A model (e.g., a ) can be trained to predict cell phenotypes (e.g., cell growth, cell death) based on the outputs of the mechanistic model, which include changes in protein localization, phosphorylation, and downstream effects
📊 Mixed-Effects Models
machine-learning
A statistical framework that incorporates both fixed effects (parameters that apply to an entire population) and random effects (parameters specific to individual groups or subjects within the data). These models are particularly valuable for analyzing hierarchical, nested, or longitudinal data where observations are not fully independent. Mixed-effects models account for both within-group and between-group variations, making them powerful tools for fields like medicine, ecology, and social sciences where data often has complex, multi-level structures.
By simultaneously modeling group-level and individual-level effects, these models provide more accurate parameter estimates and standard errors than traditional regression approaches when dealing with clustered data. They're especially useful for repeated measures designs, panel data, and any scenario where measurements are taken from the same subjects over time or under different conditions.
Example:
A mixed-effects model could be used to predict the progression of Alzheimer's disease by analyzing longitudinal MRI data, incorporating patient-specific factors and overall trends.
By simultaneously modeling group-level and individual-level effects, these models provide more accurate parameter estimates and standard errors than traditional regression approaches when dealing with clustered data. They're especially useful for repeated measures designs, panel data, and any scenario where measurements are taken from the same subjects over time or under different conditions.
Example:
A mixed-effects model could be used to predict the progression of Alzheimer's disease by analyzing longitudinal MRI data, incorporating patient-specific factors and overall trends.
🧬 Moiety
biology
Refers to a distinct part or portion of a molecule that has characteristic chemical properties and can be identified in other molecules as well. In biochemistry and organic chemistry, moieties are typically larger structural units that may contain functional groups. In pharmacology, an active moiety is the part of a molecule responsible for the physiological or pharmacological action of a drug.
📈 Monotonic
dictionary
Describing a sequence, function, or process that consistently moves in one direction without reversing. In mathematics, a monotonic function is either entirely non-decreasing (monotonically increasing) or entirely non-increasing (monotonically decreasing). For example, a monotonically increasing function satisfies f(x₁) ≤ f(x₂) whenever x₁ ≤ x₂.
In computer science and algorithms, monotonic properties are valuable for optimization and ensuring predictable behavior. Monotonic sequences are also important in analysis and calculus, where they guarantee convergence under certain conditions.
The term derives from Greek "monos" (single) and "tonos" (tone), literally meaning "single tone" or unchanging in direction.
In computer science and algorithms, monotonic properties are valuable for optimization and ensuring predictable behavior. Monotonic sequences are also important in analysis and calculus, where they guarantee convergence under certain conditions.
The term derives from Greek "monos" (single) and "tonos" (tone), literally meaning "single tone" or unchanging in direction.
🔄 Monovalent Molecular Glue Degraders
biologylab-techniques
• Definition: Small, monofunctional molecules (`<500 Da`) that induce or stabilize protein-protein interactions between an E3 ubiquitin ligase and a target protein, leading to ubiquitination and subsequent proteasomal degradation of the target.
• Mechanism of action:
- Surface modification: Alter the surface of E3 ligases to create binding interfaces for target proteins
- Neo-substrate recruitment: Induce recognition of proteins not normally targeted by the E3 ligase
- Ternary complex formation: Stabilize the interaction between E3 ligase and target protein
- Catalytic activity: Function in a substoichiometric manner to degrade multiple copies of target proteins
• Types of molecular glues:
- Type I: Induce non-native protein-protein interactions
- Type II: Stabilize protein-protein interactions
• Advantages over PROTACs:
- Lower molecular weight: Typically `<500 Da` compared to `>700` Da for PROTACs
- Improved drug-like properties: Better cell permeability and bioavailability
- Simpler chemical structure: Easier synthesis and optimization
- Potential for oral administration: Better pharmacokinetic properties
• Notable examples:
- Immunomodulatory imide drugs (IMiDs): Thalidomide, lenalidomide, pomalidomide
- Indisulam: Targets RBM39 via DCAF15
- CR8: Degrades cyclin K
- Sulfonamides: Target RBM39 through DCAF15
• Clinical significance:
- FDA-approved drugs: Thalidomide, lenalidomide, and pomalidomide for multiple myeloma
- Clinical trials: Multiple candidates in development for various cancers
- Potential for targeting previously undruggable proteins
- Emerging approach for precision medicine
• Mechanism of action:
- Surface modification: Alter the surface of E3 ligases to create binding interfaces for target proteins
- Neo-substrate recruitment: Induce recognition of proteins not normally targeted by the E3 ligase
- Ternary complex formation: Stabilize the interaction between E3 ligase and target protein
- Catalytic activity: Function in a substoichiometric manner to degrade multiple copies of target proteins
• Types of molecular glues:
- Type I: Induce non-native protein-protein interactions
- Type II: Stabilize protein-protein interactions
• Advantages over PROTACs:
- Lower molecular weight: Typically `<500 Da` compared to `>700` Da for PROTACs
- Improved drug-like properties: Better cell permeability and bioavailability
- Simpler chemical structure: Easier synthesis and optimization
- Potential for oral administration: Better pharmacokinetic properties
• Notable examples:
- Immunomodulatory imide drugs (IMiDs): Thalidomide, lenalidomide, pomalidomide
- Indisulam: Targets RBM39 via DCAF15
- CR8: Degrades cyclin K
- Sulfonamides: Target RBM39 through DCAF15
• Clinical significance:
- FDA-approved drugs: Thalidomide, lenalidomide, and pomalidomide for multiple myeloma
- Clinical trials: Multiple candidates in development for various cancers
- Potential for targeting previously undruggable proteins
- Emerging approach for precision medicine
🧩 Mosaicism Detection
biologybioinformatics
• Definition: Identification of genetic variations present in only a subset of cells within an individual, resulting from post-zygotic mutations during development.
• Types of mosaicism:
- Somatic mosaicism: Mutations present in somatic cells but not germline
- Gonadal mosaicism: Mutations present in germ cells that can be transmitted to offspring
- Chromosomal mosaicism: Presence of cells with different chromosomal compositions
- Mitochondrial heteroplasmy: Varying proportions of mutant mitochondrial DNA
• Detection methods:
- Deep sequencing: High-depth targeted sequencing to detect low-frequency variants
- Single-cell sequencing: Analyzing genetic material from individual cells
- Digital PCR: Highly sensitive detection of rare variants
- SNP arrays: Detection of mosaic copy number variations and loss of heterozygosity
• Bioinformatic challenges:
- Distinguishing true mosaic variants from sequencing errors
- Determining variant allele frequency thresholds
- Computational efficiency for large-scale analyses
- Integration of multiple data types for comprehensive detection
• Clinical significance:
- Cancer: Tumor heterogeneity and clonal evolution
- Developmental disorders: Explaining variable phenotypes
- Aging: Accumulation of throughout life
- Precision medicine: Tailoring treatments based on subclonal genetic profiles
• Types of mosaicism:
- Somatic mosaicism: Mutations present in somatic cells but not germline
- Gonadal mosaicism: Mutations present in germ cells that can be transmitted to offspring
- Chromosomal mosaicism: Presence of cells with different chromosomal compositions
- Mitochondrial heteroplasmy: Varying proportions of mutant mitochondrial DNA
• Detection methods:
- Deep sequencing: High-depth targeted sequencing to detect low-frequency variants
- Single-cell sequencing: Analyzing genetic material from individual cells
- Digital PCR: Highly sensitive detection of rare variants
- SNP arrays: Detection of mosaic copy number variations and loss of heterozygosity
• Bioinformatic challenges:
- Distinguishing true mosaic variants from sequencing errors
- Determining variant allele frequency thresholds
- Computational efficiency for large-scale analyses
- Integration of multiple data types for comprehensive detection
• Clinical significance:
- Cancer: Tumor heterogeneity and clonal evolution
- Developmental disorders: Explaining variable phenotypes
- Aging: Accumulation of throughout life
- Precision medicine: Tailoring treatments based on subclonal genetic profiles
🔗 Multi-Layer Perceptron (MLP)
machine-learning
A fundamental type of feedforward consisting of multiple layers of interconnected nodes (perceptrons). An MLP typically includes an input layer, one or more hidden layers, and an output layer, with each layer fully connected to the next. Unlike single-layer perceptrons, MLPs can learn non-linear relationships through their hidden layers and activation functions, making them capable of solving complex classification and regression problems. MLPs are the building blocks of and serve as the foundation for more sophisticated architectures like CNNs and RNNs. They're widely used in applications ranging from image recognition to financial modeling.
🩸 Myelodysplastic Syndrome (MDS)
biology
• Definition: A heterogeneous group of clonal hematopoietic stem cell disorders characterized by dysplasia and ineffective hematopoiesis in the bone marrow, leading to peripheral blood cytopenias and a variable risk of progression to acute myeloid leukemia (AML).
• Classification systems:
- World Health Organization (WHO) classification: Based on morphology, blast percentage, cytogenetics, and specific genetic abnormalities
- French-American-British (FAB) classification: Historical system based on morphological features
- International Prognostic Scoring System (IPSS) and revised IPSS (R-IPSS): Risk stratification tools that guide treatment decisions
• Molecular characteristics:
- Recurrent in genes involved in RNA splicing (SF3B1, SRSF2, U2AF1)
- Epigenetic regulators (TET2, DNMT3A, IDH1/2, ASXL1)
- Transcription factors (RUNX1, ETV6)
- Tumor suppressor genes (TP53)
- Chromosomal abnormalities (del(5q), del(7q), trisomy 8, complex karyotype)
• Clinical manifestations:
- Anemia: Fatigue, weakness, pallor, dyspnea
- Neutropenia: Increased susceptibility to infections
- Thrombocytopenia: Bleeding, bruising, petechiae
- Autoimmune phenomena in 10-20% of patients
- Potential transformation to acute myeloid leukemia
• Diagnostic approach:
- Peripheral blood smear: Cytopenias, dysplastic features in blood cells
- Bone marrow aspiration and biopsy: Dysplasia, blast percentage, cellularity
- Cytogenetic analysis: Chromosomal abnormalities
- Next-generation sequencing: Detection of
- Flow cytometry: Immunophenotypic abnormalities
• Bioinformatic applications:
- Mutational profiling to identify driver mutations and clonal architecture
- Prediction of disease progression and treatment response
- Integration of multi-omics data (genomics, transcriptomics, epigenomics)
- Development of algorithms for risk stratification
- Identification of novel therapeutic targets through pathway analysis
• Clinical significance:
- Prognosis varies widely based on subtype, cytogenetics, and molecular features
- Treatment approaches range from supportive care to targeted therapies and stem cell transplantation
- Molecular features increasingly guide personalized treatment decisions
- Model disease for studying clonal evolution and pre-leukemic states
- Emerging role of genetic predisposition in disease development
• Classification systems:
- World Health Organization (WHO) classification: Based on morphology, blast percentage, cytogenetics, and specific genetic abnormalities
- French-American-British (FAB) classification: Historical system based on morphological features
- International Prognostic Scoring System (IPSS) and revised IPSS (R-IPSS): Risk stratification tools that guide treatment decisions
• Molecular characteristics:
- Recurrent in genes involved in RNA splicing (SF3B1, SRSF2, U2AF1)
- Epigenetic regulators (TET2, DNMT3A, IDH1/2, ASXL1)
- Transcription factors (RUNX1, ETV6)
- Tumor suppressor genes (TP53)
- Chromosomal abnormalities (del(5q), del(7q), trisomy 8, complex karyotype)
• Clinical manifestations:
- Anemia: Fatigue, weakness, pallor, dyspnea
- Neutropenia: Increased susceptibility to infections
- Thrombocytopenia: Bleeding, bruising, petechiae
- Autoimmune phenomena in 10-20% of patients
- Potential transformation to acute myeloid leukemia
• Diagnostic approach:
- Peripheral blood smear: Cytopenias, dysplastic features in blood cells
- Bone marrow aspiration and biopsy: Dysplasia, blast percentage, cellularity
- Cytogenetic analysis: Chromosomal abnormalities
- Next-generation sequencing: Detection of
- Flow cytometry: Immunophenotypic abnormalities
• Bioinformatic applications:
- Mutational profiling to identify driver mutations and clonal architecture
- Prediction of disease progression and treatment response
- Integration of multi-omics data (genomics, transcriptomics, epigenomics)
- Development of algorithms for risk stratification
- Identification of novel therapeutic targets through pathway analysis
• Clinical significance:
- Prognosis varies widely based on subtype, cytogenetics, and molecular features
- Treatment approaches range from supportive care to targeted therapies and stem cell transplantation
- Molecular features increasingly guide personalized treatment decisions
- Model disease for studying clonal evolution and pre-leukemic states
- Emerging role of genetic predisposition in disease development
🧠 Neural Network
machine-learning
A computing system inspired by biological neural networks in human brains. It consists of interconnected nodes (neurons) that process and transmit information. Neural networks are fundamental to modern AI, enabling systems to learn patterns from data and make predictions or decisions.
🟢 Node.js
computer science
A JavaScript runtime environment built on Chrome's V8 JavaScript engine that allows developers to run JavaScript code outside of web browsers. Created by Ryan Dahl in 2009, Node.js enables server-side JavaScript development and has become fundamental to modern web development.
Key characteristics include:
Event-Driven Architecture: Uses an event loop for handling asynchronous operations
Non-blocking I/O: Efficiently handles multiple concurrent operations
NPM Ecosystem: Access to the world's largest package repository
Cross-Platform: Runs on Windows, macOS, and Linux
Single-Threaded: Uses one main thread with worker threads for CPU-intensive tasks
Fast Execution: Leverages V8's just-in-time compilation
Node.js is widely used for building web servers, APIs, real-time applications, microservices, and command-line tools. Its unified JavaScript environment allows developers to use the same language for both frontend and backend development.
Key characteristics include:
Event-Driven Architecture: Uses an event loop for handling asynchronous operations
Non-blocking I/O: Efficiently handles multiple concurrent operations
NPM Ecosystem: Access to the world's largest package repository
Cross-Platform: Runs on Windows, macOS, and Linux
Single-Threaded: Uses one main thread with worker threads for CPU-intensive tasks
Fast Execution: Leverages V8's just-in-time compilation
Node.js is widely used for building web servers, APIs, real-time applications, microservices, and command-line tools. Its unified JavaScript environment allows developers to use the same language for both frontend and backend development.
📈 Non-parametric Statistics
math
Statistical techniques that don't rely on assumptions about the underlying population distribution. These methods are distribution-free and typically based on ranks or orders rather than the actual values. Examples include the Mann-Whitney U test, Kruskal-Wallis test, and Spearman's rank correlation.
Non-parametric methods are more robust and flexible, making them suitable when data doesn't meet parametric assumptions or when working with ordinal data, but they may have less statistical power when parametric assumptions are valid.
Non-parametric methods are more robust and flexible, making them suitable when data doesn't meet parametric assumptions or when working with ordinal data, but they may have less statistical power when parametric assumptions are valid.
🔍⏱️ NP (Nondeterministic Polynomial Time)
computer sciencemath
NP is the complexity class of decision problems for which a solution can be verified in polynomial time, even if finding that solution might take longer. More formally, these are problems solvable by a nondeterministic Turing machine in polynomial time. Every problem in P is also in NP (since if you can solve a problem quickly, you can certainly verify a solution quickly), but the famous open question in computer science is whether P = NP, which asks if every problem whose solution can be quickly verified can also be quickly solved. Examples of NP problems include the Boolean satisfiability problem, the traveling salesman decision problem, and the subset sum problem.
🧩🔄 NP-Complete
computer sciencemath
NP-Complete problems are the hardest problems in the NP class, in the sense that if an efficient (polynomial time) algorithm exists for any NP-Complete problem, then efficient algorithms would exist for all problems in NP. A problem is NP-Complete if it is in NP and every other problem in NP can be reduced to it in polynomial time. The first problem proven to be NP-Complete was the Boolean satisfiability problem (SAT), through Cook's theorem. Other examples include the traveling salesman decision problem, the graph coloring problem, and the subset sum problem. The P vs NP question essentially asks whether NP-Complete problems can be solved efficiently.
🏋️♂️🧠 NP-Hard
computer sciencemath
NP-Hard problems are at least as hard as the hardest problems in NP, but they might not be in NP themselves. A problem is NP-Hard if every problem in NP can be reduced to it in polynomial time, but the problem itself might not be verifiable in polynomial time. NP-Hard problems can be decision problems, search problems, or optimization problems. All problems are NP-Hard, but not all NP-Hard problems are . Examples of NP-Hard problems include the traveling salesman optimization problem (finding the shortest route), the graph isomorphism problem, and the halting problem. Many important optimization problems in various fields like operations research, bioinformatics, and artificial intelligence are NP-Hard, which is why approximation algorithms and heuristics are often used to find good-enough solutions in practice.
⚡🎯 Optimizer
machine-learningmath
An algorithm used to adjust the parameters of models during training to minimize the . Optimizers determine how the model's weights and biases are updated based on the computed gradients, directly affecting the speed and quality of learning.
Common optimizers include:
** (SGD)**: The fundamental optimizer that updates parameters in the direction opposite to the gradient.
Adam (Adaptive Moment Estimation): Combines momentum and adaptive learning rates, widely used for its robustness and efficiency.
RMSprop: Adapts learning rates based on recent gradient magnitudes, effective for non-stationary objectives.
AdaGrad: Adapts learning rates based on historical gradients, useful for sparse data.
Momentum: Accelerates SGD by adding a fraction of the previous update to the current one.
AdamW: A variant of Adam with decoupled weight decay for better regularization.
The choice of optimizer significantly impacts training convergence, stability, and final model performance. Modern frameworks typically default to Adam or its variants due to their adaptive nature and robust performance across various tasks.
Common optimizers include:
** (SGD)**: The fundamental optimizer that updates parameters in the direction opposite to the gradient.
Adam (Adaptive Moment Estimation): Combines momentum and adaptive learning rates, widely used for its robustness and efficiency.
RMSprop: Adapts learning rates based on recent gradient magnitudes, effective for non-stationary objectives.
AdaGrad: Adapts learning rates based on historical gradients, useful for sparse data.
Momentum: Accelerates SGD by adding a fraction of the previous update to the current one.
AdamW: A variant of Adam with decoupled weight decay for better regularization.
The choice of optimizer significantly impacts training convergence, stability, and final model performance. Modern frameworks typically default to Adam or its variants due to their adaptive nature and robust performance across various tasks.
🧬 Orthologs
biology
• Definition: Genes in different species that evolved from a common ancestral gene following a speciation event.
• Key characteristics:
- Typically maintain similar functions across species
- Sequence similarity reflects evolutionary distance between species
- Usually present as single copies in each genome (except after lineage-specific duplications)
- Conserved genomic context (synteny) in closely related species
- Often used as evolutionary markers for phylogenetic studies
• Evolutionary significance:
- Enable comparative genomic studies across species
- Provide insights into gene function conservation and divergence
- Allow reconstruction of species phylogenies
- Help identify core genes essential for life
• Identification methods:
- Sequence similarity: BLAST, HMMER, and other alignment tools
- Phylogenetic analysis: Tree-based methods to distinguish orthologs from
- Synteny analysis: Examining conservation of gene order and chromosomal context
- Reciprocal best hits (RBH): Identifying mutual best matches between genomes
• Applications in bioinformatics:
- Functional annotation: Transferring knowledge from well-studied to newly sequenced organisms
- Comparative genomics: Understanding genome evolution and species relationships
- Identification of conserved regulatory elements
- Drug target discovery across model organisms
• Key characteristics:
- Typically maintain similar functions across species
- Sequence similarity reflects evolutionary distance between species
- Usually present as single copies in each genome (except after lineage-specific duplications)
- Conserved genomic context (synteny) in closely related species
- Often used as evolutionary markers for phylogenetic studies
• Evolutionary significance:
- Enable comparative genomic studies across species
- Provide insights into gene function conservation and divergence
- Allow reconstruction of species phylogenies
- Help identify core genes essential for life
• Identification methods:
- Sequence similarity: BLAST, HMMER, and other alignment tools
- Phylogenetic analysis: Tree-based methods to distinguish orthologs from
- Synteny analysis: Examining conservation of gene order and chromosomal context
- Reciprocal best hits (RBH): Identifying mutual best matches between genomes
• Applications in bioinformatics:
- Functional annotation: Transferring knowledge from well-studied to newly sequenced organisms
- Comparative genomics: Understanding genome evolution and species relationships
- Identification of conserved regulatory elements
- Drug target discovery across model organisms
📈 Overfitting
machine-learning
A common problem in where a model learns the training data too precisely, including its noise and irregularities. This reduces its ability to generalize to new data, like memorizing answers instead of understanding the underlying concepts.
⏱️✓ P (Polynomial Time)
computer sciencemath
P is the complexity class of decision problems that can be solved by a deterministic Turing machine in polynomial time. In simpler terms, these are problems for which efficient algorithms exist that can find a solution in a reasonable amount of time, even as the input size grows. The time required to solve these problems grows as a polynomial function of the input size. Examples include sorting algorithms, searching in ordered lists, and determining if a number is prime using modern primality tests. P represents the class of problems that are considered computationally tractable.
🧬 Paralogs
biology
• Definition: Genes within the same organism that arose from gene duplication events and have subsequently diverged in function.
• Key characteristics:
- Share sequence similarity due to common ancestry
- Often develop distinct but related functions (subfunctionalization or neofunctionalization)
- May exhibit different expression patterns across tissues or developmental stages
- Can have varying degrees of sequence conservation depending on duplication age
• Evolutionary mechanisms:
- Whole genome duplication (polyploidization)
- Segmental duplication of chromosomal regions
- Tandem duplication of individual genes
- Retrotransposition (RNA-mediated duplication)
• Biological significance:
- Source of genetic redundancy providing robustness to mutations
- Platform for evolutionary innovation through functional divergence
- Contribute to species-specific adaptations and phenotypic diversity
- Allow specialization of protein functions for different cellular contexts
• Examples in humans:
- Hemoglobin subunits (α, β, γ, δ)
- Myosin heavy chain
- Hox gene clusters
- Olfactory receptor gene family
• Applications in bioinformatics:
- Phylogenetic analysis to determine duplication timing
- Functional prediction of uncharacterized genes
- Identification of lineage-specific gene expansions
- Drug target discovery and specificity assessment
• Key characteristics:
- Share sequence similarity due to common ancestry
- Often develop distinct but related functions (subfunctionalization or neofunctionalization)
- May exhibit different expression patterns across tissues or developmental stages
- Can have varying degrees of sequence conservation depending on duplication age
• Evolutionary mechanisms:
- Whole genome duplication (polyploidization)
- Segmental duplication of chromosomal regions
- Tandem duplication of individual genes
- Retrotransposition (RNA-mediated duplication)
• Biological significance:
- Source of genetic redundancy providing robustness to mutations
- Platform for evolutionary innovation through functional divergence
- Contribute to species-specific adaptations and phenotypic diversity
- Allow specialization of protein functions for different cellular contexts
• Examples in humans:
- Hemoglobin subunits (α, β, γ, δ)
- Myosin heavy chain
- Hox gene clusters
- Olfactory receptor gene family
• Applications in bioinformatics:
- Phylogenetic analysis to determine duplication timing
- Functional prediction of uncharacterized genes
- Identification of lineage-specific gene expansions
- Drug target discovery and specificity assessment
📊 Parametric Statistics
math
Statistical methods that assume data comes from a population following a probability distribution based on a fixed set of parameters. These methods make specific assumptions about the data's distribution (often assuming normal distribution) and draw inferences about the parameters of the assumed distribution. Examples include t-tests, ANOVA, and linear regression.
Parametric methods are generally more powerful when their assumptions are met but may produce misleading results when these assumptions are violated.
Parametric methods are generally more powerful when their assumptions are met but may produce misleading results when these assumptions are violated.
📊 Pearson Correlation Coefficient
mathmachine-learning
The Pearson Correlation Coefficient (PCC) is a statistical measure that quantifies the linear relationship between two continuous variables. It produces a value ranging from -1 to +1, where +1 indicates a perfect positive linear relationship, -1 indicates a perfect negative linear relationship, and 0 indicates no linear relationship between the variables.
Mathematically, it is calculated as the ratio between the covariance of two variables and the product of their standard deviations, making it a normalized measurement of covariance. The formula is often expressed as:
$$
r = \frac{\sum_{i=1}^{n} (x_i - \bar{x})(y_i - \bar{y})}{\sqrt{\sum_{i=1}^{n} (x_i - \bar{x})^2 \sum_{i=1}^{n} (y_i - \bar{y})^2}}
$$
In , the Pearson correlation coefficient serves several critical functions:
1. Feature Selection: It helps identify which features have strong relationships with the target variable, allowing data scientists to select the most relevant features for model training.
2. Multicollinearity Detection: It identifies highly correlated input features that might cause instability in models like linear regression.
3. Dimensionality Reduction: Understanding correlation patterns helps in techniques like Principal Component Analysis (PCA) to reduce the number of features while preserving information.
4. Data Exploration: It provides insights into relationships within the data, guiding further analysis and model selection.
The interpretation of correlation strength varies by field, but generally:
- Values between ±0.1 and ±0.3 indicate weak correlation
- Values between ±0.3 and ±0.5 indicate moderate correlation
- Values between ±0.5 and ±1.0 indicate strong correlation
It's important to note that Pearson correlation only captures linear relationships and is sensitive to outliers. For non-linear relationships or when dealing with ordinal data, alternative measures like Spearman's rank correlation coefficient may be more appropriate.
In practical applications, Pearson correlation is used in genomics to identify relationships between genes, in financial modeling to analyze market dependencies, and in recommendation systems to measure similarities between user preferences or items.
Simple Examples:
1. Strong Positive Correlation (r ≈ 0.9): Height and weight in a population. As height increases, weight tends to increase proportionally.
2. Moderate Positive Correlation (r ≈ 0.4): Study hours and test scores. More study time generally leads to better scores, but other factors also influence performance.
3. No Correlation (r ≈ 0): Shoe size and intelligence. These variables have no meaningful linear relationship.
4. Moderate Negative Correlation (r ≈ -0.4): Age of a car and its resale value. Older cars typically have lower resale values, though condition and other factors matter.
5. Strong Negative Correlation (r ≈ -0.8): Outdoor temperature and home heating usage. As temperature drops, heating usage increases substantially.
Mathematically, it is calculated as the ratio between the covariance of two variables and the product of their standard deviations, making it a normalized measurement of covariance. The formula is often expressed as:
$$
r = \frac{\sum_{i=1}^{n} (x_i - \bar{x})(y_i - \bar{y})}{\sqrt{\sum_{i=1}^{n} (x_i - \bar{x})^2 \sum_{i=1}^{n} (y_i - \bar{y})^2}}
$$
In , the Pearson correlation coefficient serves several critical functions:
1. Feature Selection: It helps identify which features have strong relationships with the target variable, allowing data scientists to select the most relevant features for model training.
2. Multicollinearity Detection: It identifies highly correlated input features that might cause instability in models like linear regression.
3. Dimensionality Reduction: Understanding correlation patterns helps in techniques like Principal Component Analysis (PCA) to reduce the number of features while preserving information.
4. Data Exploration: It provides insights into relationships within the data, guiding further analysis and model selection.
The interpretation of correlation strength varies by field, but generally:
- Values between ±0.1 and ±0.3 indicate weak correlation
- Values between ±0.3 and ±0.5 indicate moderate correlation
- Values between ±0.5 and ±1.0 indicate strong correlation
It's important to note that Pearson correlation only captures linear relationships and is sensitive to outliers. For non-linear relationships or when dealing with ordinal data, alternative measures like Spearman's rank correlation coefficient may be more appropriate.
In practical applications, Pearson correlation is used in genomics to identify relationships between genes, in financial modeling to analyze market dependencies, and in recommendation systems to measure similarities between user preferences or items.
Simple Examples:
1. Strong Positive Correlation (r ≈ 0.9): Height and weight in a population. As height increases, weight tends to increase proportionally.
2. Moderate Positive Correlation (r ≈ 0.4): Study hours and test scores. More study time generally leads to better scores, but other factors also influence performance.
3. No Correlation (r ≈ 0): Shoe size and intelligence. These variables have no meaningful linear relationship.
4. Moderate Negative Correlation (r ≈ -0.4): Age of a car and its resale value. Older cars typically have lower resale values, though condition and other factors matter.
5. Strong Negative Correlation (r ≈ -0.8): Outdoor temperature and home heating usage. As temperature drops, heating usage increases substantially.
🧬 Pleiotropy
biology
• Definition: The phenomenon in which a single gene or genetic variant influences multiple phenotypic traits. The term was coined by Ludwig Plate in 1910, derived from Greek words "pleíōn" (more) and "trópos" (turn, way, manner). 1
• Types of pleiotropy:
- Biological (horizontal) pleiotropy: A genetic variant directly affects multiple traits through independent biological pathways 1
- Mediated (vertical/relational) pleiotropy: A variant influences one trait, which in turn causes changes in secondary traits 1 4
- Spurious pleiotropy: Statistical or methodological biases create false associations between a variant and multiple traits 1
- Selectional pleiotropy: When a single phenotype influences evolutionary fitness in multiple ways depending on factors like age and sex 1 4
• Molecular mechanisms:
- Multiple protein interactions: A gene product interacts with multiple proteins or catalyzes different reactions 4
- : One gene produces different protein with varied structures and functions 1
- Regulatory effects: A variant alters the expression of many genes or influences chromosome 3D structure 1
- Signaling cascades: A primary gene product initiates multiple downstream effects 2
• Biological significance:
- Evolutionary constraints: Pleiotropic genes can limit the rate of multivariate evolution when selection on different traits favors different alleles 1
- Genetic disorders: Many genetic diseases exhibit pleiotropy, such as phenylketonuria (PKU) affecting both nervous and integumentary systems 1
- Development: Pleiotropic genes often play crucial roles in developmental processes and cell fate decisions 5
- Stabilizing selection: Highly pleiotropic genes typically experience strong stabilizing selection as they affect multiple phenotypes 5
• Applications in bioinformatics:
- Genome-wide association studies (GWAS): Identifying variants linked to multiple traits or diseases 1
- Multivariate analysis methods: Statistical approaches like multivariate functional linear models to analyze pleiotropic effects on quantitative traits 3
- Network biology: Mapping gene interaction networks to understand pleiotropic effects 4
- Disease gene discovery: Using pleiotropy patterns to identify novel disease genes and understand complex disorders 5
- Pharmacogenomics: Understanding how genetic variants may affect multiple drug responses or side effects
• Clinical relevance:
- Syndromic diseases: Understanding the diverse manifestations of genetic disorders affecting multiple systems 5
- Drug development: Considering pleiotropic effects when targeting specific genes to minimize unintended consequences
- Personalized medicine: Accounting for pleiotropic effects when predicting disease risk or treatment outcomes
- Aging research: Antagonistic pleiotropy model explains how genes beneficial early in life may have detrimental effects later
• Types of pleiotropy:
- Biological (horizontal) pleiotropy: A genetic variant directly affects multiple traits through independent biological pathways 1
- Mediated (vertical/relational) pleiotropy: A variant influences one trait, which in turn causes changes in secondary traits 1 4
- Spurious pleiotropy: Statistical or methodological biases create false associations between a variant and multiple traits 1
- Selectional pleiotropy: When a single phenotype influences evolutionary fitness in multiple ways depending on factors like age and sex 1 4
• Molecular mechanisms:
- Multiple protein interactions: A gene product interacts with multiple proteins or catalyzes different reactions 4
- : One gene produces different protein with varied structures and functions 1
- Regulatory effects: A variant alters the expression of many genes or influences chromosome 3D structure 1
- Signaling cascades: A primary gene product initiates multiple downstream effects 2
• Biological significance:
- Evolutionary constraints: Pleiotropic genes can limit the rate of multivariate evolution when selection on different traits favors different alleles 1
- Genetic disorders: Many genetic diseases exhibit pleiotropy, such as phenylketonuria (PKU) affecting both nervous and integumentary systems 1
- Development: Pleiotropic genes often play crucial roles in developmental processes and cell fate decisions 5
- Stabilizing selection: Highly pleiotropic genes typically experience strong stabilizing selection as they affect multiple phenotypes 5
• Applications in bioinformatics:
- Genome-wide association studies (GWAS): Identifying variants linked to multiple traits or diseases 1
- Multivariate analysis methods: Statistical approaches like multivariate functional linear models to analyze pleiotropic effects on quantitative traits 3
- Network biology: Mapping gene interaction networks to understand pleiotropic effects 4
- Disease gene discovery: Using pleiotropy patterns to identify novel disease genes and understand complex disorders 5
- Pharmacogenomics: Understanding how genetic variants may affect multiple drug responses or side effects
• Clinical relevance:
- Syndromic diseases: Understanding the diverse manifestations of genetic disorders affecting multiple systems 5
- Drug development: Considering pleiotropic effects when targeting specific genes to minimize unintended consequences
- Personalized medicine: Accounting for pleiotropic effects when predicting disease risk or treatment outcomes
- Aging research: Antagonistic pleiotropy model explains how genes beneficial early in life may have detrimental effects later
🔄 Pluripotency
biology
• Definition: The ability of stem cells to differentiate into any cell type derived from the three germ layers (endoderm, mesoderm, and ectoderm) that form during embryonic development.
• Types of pluripotent cells:
- Embryonic stem cells (ESCs): Derived from the inner cell mass of blastocysts
- Induced pluripotent stem cells (iPSCs): Somatic cells reprogrammed to a pluripotent state
- Embryonic germ cells: Derived from primordial germ cells
- Embryonal carcinoma cells: Derived from teratocarcinomas
• Molecular mechanisms:
- Core transcription factor network: OCT4, SOX2, and NANOG maintain the pluripotent state
- Epigenetic regulation: DNA methylation, histone modifications, and
- Signaling pathways: LIF/STAT3, Wnt/β-catenin, TGF-β/Activin/Nodal, and FGF pathways
- MicroRNAs: Regulation of gene expression at the post-transcriptional level
• Applications in research:
- Disease modeling: Creating patient-specific cell lines to study disease mechanisms
- Drug discovery and toxicity testing: Screening compounds on relevant human cell types
- Developmental biology: Understanding cellular differentiation and embryonic development
- Regenerative medicine: Source for cell replacement therapies
• Clinical relevance:
- Regenerative medicine: Potential treatments for degenerative diseases and injuries
- Cancer biology: Understanding similarities between pluripotency and oncogenic transformation
- Aging research: Insights into cellular rejuvenation and senescence
- Ethical considerations: Debates surrounding embryonic stem cell research and therapeutic cloning
• Types of pluripotent cells:
- Embryonic stem cells (ESCs): Derived from the inner cell mass of blastocysts
- Induced pluripotent stem cells (iPSCs): Somatic cells reprogrammed to a pluripotent state
- Embryonic germ cells: Derived from primordial germ cells
- Embryonal carcinoma cells: Derived from teratocarcinomas
• Molecular mechanisms:
- Core transcription factor network: OCT4, SOX2, and NANOG maintain the pluripotent state
- Epigenetic regulation: DNA methylation, histone modifications, and
- Signaling pathways: LIF/STAT3, Wnt/β-catenin, TGF-β/Activin/Nodal, and FGF pathways
- MicroRNAs: Regulation of gene expression at the post-transcriptional level
• Applications in research:
- Disease modeling: Creating patient-specific cell lines to study disease mechanisms
- Drug discovery and toxicity testing: Screening compounds on relevant human cell types
- Developmental biology: Understanding cellular differentiation and embryonic development
- Regenerative medicine: Source for cell replacement therapies
• Clinical relevance:
- Regenerative medicine: Potential treatments for degenerative diseases and injuries
- Cancer biology: Understanding similarities between pluripotency and oncogenic transformation
- Aging research: Insights into cellular rejuvenation and senescence
- Ethical considerations: Debates surrounding embryonic stem cell research and therapeutic cloning
🧮 Pohlig-Hellman Algorithm
computer sciencemath
An algorithm for computing discrete logarithms in a cyclic group, particularly useful when the order of the group has only small prime factors. Developed by Stephen Pohlig and Martin Hellman in 1978, it significantly reduces the computational complexity of the discrete logarithm problem in certain groups.
The algorithm works by decomposing the discrete logarithm problem in a group of composite order into smaller subproblems in groups of prime order using the . It then solves these smaller problems using techniques like the baby-step giant-step algorithm. While highly efficient for groups whose order factors into small primes, it's ineffective against groups specifically chosen for cryptographic purposes (those with at least one large prime factor). Understanding this algorithm is crucial for cryptographers to select appropriate parameters for discrete logarithm-based cryptosystems like Diffie-Hellman and ElGamal.
Video explanation:
How can I compute discrete logs faster? — Pohlig–Hellman — The Ross Program - A mathematical explanation of how the Diffie-Hellman protocol works and why it's secure.
The algorithm works by decomposing the discrete logarithm problem in a group of composite order into smaller subproblems in groups of prime order using the . It then solves these smaller problems using techniques like the baby-step giant-step algorithm. While highly efficient for groups whose order factors into small primes, it's ineffective against groups specifically chosen for cryptographic purposes (those with at least one large prime factor). Understanding this algorithm is crucial for cryptographers to select appropriate parameters for discrete logarithm-based cryptosystems like Diffie-Hellman and ElGamal.
Video explanation:
How can I compute discrete logs faster? — Pohlig–Hellman — The Ross Program - A mathematical explanation of how the Diffie-Hellman protocol works and why it's secure.
🚪 Ports
networkingsystem-administration
Numerical identifiers (0-65535) that specify which service or application should handle network communication on a server. Ports act like doors or channels that allow multiple network services to run simultaneously on the same computer.
Ports are specified in URLs after the name, separated by a colon (e.g., example.com:8080). If no port is specified, default ports are used based on the protocol.
Common Default Ports:
- 80: HTTP (web traffic)
- 443: HTTPS (secure web traffic)
- 21: FTP (file transfer)
- 22: SSH (secure shell)
- 25: SMTP (email sending)
- 53: DNS ( name resolution)
- 3306: MySQL database
- 5432: PostgreSQL database
- 8080: Alternative HTTP (often for development)
Port Categories:
- Well-known ports (0-1023): Reserved for system services
- Registered ports (1024-49151): Assigned to specific applications
- Dynamic/Private ports (49152-65535): Available for general use
Ports enable network traffic routing and allow servers to host multiple services simultaneously, such as a web server on port 80 and an email server on port 25.
Ports are specified in URLs after the name, separated by a colon (e.g., example.com:8080). If no port is specified, default ports are used based on the protocol.
Common Default Ports:
- 80: HTTP (web traffic)
- 443: HTTPS (secure web traffic)
- 21: FTP (file transfer)
- 22: SSH (secure shell)
- 25: SMTP (email sending)
- 53: DNS ( name resolution)
- 3306: MySQL database
- 5432: PostgreSQL database
- 8080: Alternative HTTP (often for development)
Port Categories:
- Well-known ports (0-1023): Reserved for system services
- Registered ports (1024-49151): Assigned to specific applications
- Dynamic/Private ports (49152-65535): Available for general use
Ports enable network traffic routing and allow servers to host multiple services simultaneously, such as a web server on port 80 and an email server on port 25.
📍 Positional Encoding
machine-learning
A technique that adds location information to data in , helping them understand the order and structure of sequences. It's crucial for models to process ordered data like text or time series, as it provides context about where each piece of information belongs in the sequence.
⌨️ Prompt Engineering
machine-learning
The art and science of crafting effective inputs for large language models to achieve desired outputs. It involves understanding model behavior and using specific techniques to guide the model's responses, crucial for getting optimal results from AI systems.
🧬 PROteolysis TArgeting Chimeras (PROTACs)
biologylab-techniques
• Definition: Bifunctional molecules designed to induce by simultaneously binding to a protein of interest and an E3 ubiquitin ligase, bringing them into proximity to facilitate ubiquitination and subsequent proteasomal degradation of the target protein.
• Structural components:
- Target protein-binding ligand: Binds specifically to the protein of interest
- E3 ligase-binding ligand: Recruits an E3 ubiquitin ligase (e.g., CRBN, VHL, IAP, MDM2)
- Linker: Connects the two ligands and optimizes their spatial arrangement
• Mechanism of action:
- Target engagement: Binding to both the target protein and E3 ligase
- Ternary complex formation: Creation of a three-molecule complex (target-PROTAC-E3 ligase)
- Ubiquitination: Transfer of ubiquitin molecules to the target protein
- Proteasomal degradation: Recognition and degradation of the polyubiquitinated target
- Recycling: Release of the PROTAC for additional rounds of target degradation
• Advantages over traditional inhibitors:
- Event-driven pharmacology: Effect persists after drug clearance
- Catalytic mechanism: One PROTAC can facilitate degradation of multiple target proteins
- Broader target scope: Can address previously undruggable proteins
- Potential to overcome resistance: Complete protein removal versus functional inhibition
- Degradation of all protein functions: Not limited to active site inhibition
• Design considerations:
- E3 ligase selection: Tissue expression, binding affinity, and substrate compatibility
- Linker optimization: Length, composition, and flexibility
- Target ligand selection: Binding affinity, selectivity, and attachment point
- Ternary complex geometry: Spatial arrangement for optimal ubiquitination
• Clinical development status:
- Multiple candidates in clinical trials for various cancers
- ARV-110 (androgen receptor degrader) for prostate cancer
- ARV-471 (estrogen receptor degrader) for breast cancer
- DT2216 (BCL-xL degrader) for hematologic malignancies
• Challenges and limitations:
- High molecular weight: Potential issues with cell permeability and oral bioavailability
- Complex structure: Synthetic challenges and potential metabolic instability
- Hook effect: Decreased efficacy at high concentrations
- Tissue-specific E3 ligase expression: Potential limitations in tissue selectivity
• Structural components:
- Target protein-binding ligand: Binds specifically to the protein of interest
- E3 ligase-binding ligand: Recruits an E3 ubiquitin ligase (e.g., CRBN, VHL, IAP, MDM2)
- Linker: Connects the two ligands and optimizes their spatial arrangement
• Mechanism of action:
- Target engagement: Binding to both the target protein and E3 ligase
- Ternary complex formation: Creation of a three-molecule complex (target-PROTAC-E3 ligase)
- Ubiquitination: Transfer of ubiquitin molecules to the target protein
- Proteasomal degradation: Recognition and degradation of the polyubiquitinated target
- Recycling: Release of the PROTAC for additional rounds of target degradation
• Advantages over traditional inhibitors:
- Event-driven pharmacology: Effect persists after drug clearance
- Catalytic mechanism: One PROTAC can facilitate degradation of multiple target proteins
- Broader target scope: Can address previously undruggable proteins
- Potential to overcome resistance: Complete protein removal versus functional inhibition
- Degradation of all protein functions: Not limited to active site inhibition
• Design considerations:
- E3 ligase selection: Tissue expression, binding affinity, and substrate compatibility
- Linker optimization: Length, composition, and flexibility
- Target ligand selection: Binding affinity, selectivity, and attachment point
- Ternary complex geometry: Spatial arrangement for optimal ubiquitination
• Clinical development status:
- Multiple candidates in clinical trials for various cancers
- ARV-110 (androgen receptor degrader) for prostate cancer
- ARV-471 (estrogen receptor degrader) for breast cancer
- DT2216 (BCL-xL degrader) for hematologic malignancies
• Challenges and limitations:
- High molecular weight: Potential issues with cell permeability and oral bioavailability
- Complex structure: Synthetic challenges and potential metabolic instability
- Hook effect: Decreased efficacy at high concentrations
- Tissue-specific E3 ligase expression: Potential limitations in tissue selectivity
🔗 Protocol/Scheme
networkingweb-development
The first part of a URL that specifies the communication protocol or method used to access a resource on the internet. Common protocols include HTTP (Hypertext Transfer Protocol), HTTPS (HTTP Secure), FTP (File Transfer Protocol), and others.
The protocol/scheme appears at the beginning of a URL followed by a colon and two forward slashes (://). For example, in 'https://www.example.com', 'https' is the protocol that indicates secure HTTP communication should be used.
Common Protocols:
- HTTP: Standard web protocol for transferring web pages
- HTTPS: Secure version of HTTP with encryption
- FTP: File Transfer Protocol for uploading/downloading files
- SMTP: Simple Mail Transfer Protocol for email
- SSH: Secure Shell for remote server access
- FILE: Local file system access
The protocol determines how the client (browser) will communicate with the server to retrieve the requested resource.
The protocol/scheme appears at the beginning of a URL followed by a colon and two forward slashes (://). For example, in 'https://www.example.com', 'https' is the protocol that indicates secure HTTP communication should be used.
Common Protocols:
- HTTP: Standard web protocol for transferring web pages
- HTTPS: Secure version of HTTP with encryption
- FTP: File Transfer Protocol for uploading/downloading files
- SMTP: Simple Mail Transfer Protocol for email
- SSH: Secure Shell for remote server access
- FILE: Local file system access
The protocol determines how the client (browser) will communicate with the server to retrieve the requested resource.
➗ Quotient Rule
math
A differentiation rule used to find the derivative of a function that is the quotient (division) of two other functions. If you have a function h(x) = f(x)/g(x), where both f(x) and g(x) are differentiable and g(x) ≠ 0, then the quotient rule provides the formula for h'(x).
The quotient rule formula is:
Often remembered by the mnemonic "low d-high minus high d-low, over low squared" where "high" refers to the numerator function and "low" refers to the denominator function. This rule is particularly useful in calculus for differentiating rational functions, rates of change problems, and optimization problems involving ratios.
The quotient rule formula is:
Often remembered by the mnemonic "low d-high minus high d-low, over low squared" where "high" refers to the numerator function and "low" refers to the denominator function. This rule is particularly useful in calculus for differentiating rational functions, rates of change problems, and optimization problems involving ratios.
🔄 Recurrent Neural Network (RNN)
machine-learning
A designed for sequential data that maintains a "memory" of previous inputs. Like having a short-term memory, it processes information in order and uses past context to understand current inputs, making it suitable for tasks like text prediction and time series analysis.
🎮 Reinforcement Learning
machine-learning
A type of where an agent learns to make decisions by interacting with an environment. The agent receives rewards or penalties based on its actions, similar to how humans learn through trial and error. It's crucial for robotics, game AI, and autonomous systems.
⚡ ReLU (Rectified Linear Unit)
machine-learningmath
A widely-used activation function in that outputs the input directly if it's positive, otherwise it outputs zero. Mathematically defined as f(x) = max(0, x), ReLU is simple yet effective at introducing non-linearity into while being computationally efficient. It helps solve the vanishing gradient problem that plagued earlier activation functions like sigmoid and tanh, allowing for faster training of deep networks. ReLU has become the default activation function for hidden layers in most modern architectures, though variants like Leaky ReLU and ELU address some of its limitations, such as the "dying ReLU" problem where neurons can become permanently inactive.
🔄🚀 Repository Dispatch:
git
A GitHub feature that enables external events to trigger GitHub Actions workflows in a repository. It allows custom webhook events to initiate automated processes, facilitating integration with external services and enabling programmatic workflow execution. Repository dispatch is commonly used for creating custom automation triggers and coordinating workflows across multiple repositories.
🌐 RESTful API
computer sciencenetworkingweb development
An architectural style for designing web services that follows REST (Representational State Transfer) principles. RESTful APIs use standard HTTP methods to perform operations on resources identified by URLs. They are stateless, meaning each request contains all necessary information, and typically return data in JSON format.
HTTP Methods:
GET: Retrieves data from a resource without modifying it. Safe and idempotent operation used for reading data.
- Example: `GET /users/123` - Retrieve user with ID 123
POST: Creates new resources or submits data for processing. Not idempotent, as multiple requests may create multiple resources.
- Example: `POST /users` - Create a new user
PUT: Updates or replaces an entire resource. Idempotent operation that overwrites the complete resource.
- Example: `PUT /users/123` - Replace user 123 with new data
PATCH: Partially updates a resource by modifying only specified fields. More efficient than PUT for small changes.
- Example: `PATCH /users/123` - Update only specific fields of user 123
DELETE: Removes a resource from the server. Idempotent operation for resource deletion.
- Example: `DELETE /users/123` - Delete user with ID 123
HEAD: Retrieves only the headers of a resource without the body content. Useful for checking resource existence or metadata.
- Example: `HEAD /users/123` - Check if user 123 exists
Key Characteristics:
Resource-based: Everything is treated as a resource with a unique URL
Stateless: Each request is independent and contains all needed information
Cacheable: Responses can be cached to improve performance
Uniform Interface: Consistent way of interacting with resources
Layered System: Can include intermediary layers like proxies and gateways
RESTful APIs are widely used for web applications, mobile apps, and microservices due to their simplicity, scalability, and compatibility with web standards.
HTTP Methods:
GET: Retrieves data from a resource without modifying it. Safe and idempotent operation used for reading data.
- Example: `GET /users/123` - Retrieve user with ID 123
POST: Creates new resources or submits data for processing. Not idempotent, as multiple requests may create multiple resources.
- Example: `POST /users` - Create a new user
PUT: Updates or replaces an entire resource. Idempotent operation that overwrites the complete resource.
- Example: `PUT /users/123` - Replace user 123 with new data
PATCH: Partially updates a resource by modifying only specified fields. More efficient than PUT for small changes.
- Example: `PATCH /users/123` - Update only specific fields of user 123
DELETE: Removes a resource from the server. Idempotent operation for resource deletion.
- Example: `DELETE /users/123` - Delete user with ID 123
HEAD: Retrieves only the headers of a resource without the body content. Useful for checking resource existence or metadata.
- Example: `HEAD /users/123` - Check if user 123 exists
Key Characteristics:
Resource-based: Everything is treated as a resource with a unique URL
Stateless: Each request is independent and contains all needed information
Cacheable: Responses can be cached to improve performance
Uniform Interface: Consistent way of interacting with resources
Layered System: Can include intermediary layers like proxies and gateways
RESTful APIs are widely used for web applications, mobile apps, and microservices due to their simplicity, scalability, and compatibility with web standards.
📈 Ridge Regression
mathmachine-learning
A regularization technique that addresses multicollinearity in linear regression by adding an L2 penalty term to the cost function. Unlike Lasso, Ridge Regression shrinks coefficients toward zero but rarely sets them exactly to zero, keeping all features in the model while reducing their impact. This approach is particularly effective when dealing with highly correlated predictors, preventing the model from assigning excessive importance to any single variable.
Ridge Regression excels in scenarios where all features contribute to the outcome but need to be constrained to prevent , such as in economic forecasting, climate modeling, and biomedical research.
Ridge Regression excels in scenarios where all features contribute to the outcome but need to be constrained to prevent , such as in economic forecasting, climate modeling, and biomedical research.
🧬 RNA-seq (RNA sequencing)
biologylab-techniques
• Purpose: Measures gene expression — tells you which genes are being transcribed into RNA, and how much.
• Data type: Sequencing reads aligned to genes/transcripts.
• Used for: Identifying differentially expressed genes between conditions (e.g., normal vs. cancer cells).
• Data type: Sequencing reads aligned to genes/transcripts.
• Used for: Identifying differentially expressed genes between conditions (e.g., normal vs. cancer cells).
🧬 sgRNA (Single Guide RNA)
biologylab-techniques
• Definition: A synthetic RNA molecule that combines the functions of crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA) into a single structure, used to guide Cas nucleases to specific DNA targets in CRISPR-Cas genome editing systems.
• Structure and components:
- Spacer sequence (20 nucleotides): Complementary to the target DNA sequence
- Scaffold sequence (~80 nucleotides): Forms secondary structures necessary for Cas protein binding
- PAM (Protospacer Adjacent Motif): DNA sequence required for target recognition (not part of sgRNA but essential for targeting)
• Design considerations:
- Target specificity: Minimizing off-target effects through careful sequence selection
- GC content: Optimal range of 40-60% for efficient binding
- Secondary structure: Avoiding self-complementarity that could interfere with target binding
- Position effects: Targeting the beginning of genes or critical functional
- PAM proximity: Selecting targets with appropriate PAM sequences for the Cas variant used
• Bioinformatic tools for sgRNA design:
- CHOPCHOP: Web tool for CRISPR/Cas9 target prediction and off-target evaluation
- CRISPOR: Comprehensive tool for guide selection and off-target prediction
- E-CRISP: Design tool with evaluation of on-target efficiency and off-target effects
- Cas-Designer: Tool for designing guide RNAs for various CRISPR systems
- CRISPRscan: Algorithm for predicting sgRNA efficiency in vivo
• Applications in genome editing:
- Gene knockout: Introducing frameshift mutations through NHEJ repair
- Gene knock-in: Precise sequence insertion via HDR pathway
- Base editing: Creating specific nucleotide changes without double-strand breaks
- Epigenetic modification: Targeting chromatin modifiers to specific genomic loci
- Transcriptional regulation: Activating or repressing gene expression (CRISPRa/CRISPRi)
- Multiplexed editing: Simultaneous modification of multiple genomic targets
• Computational challenges:
- Off-target prediction: Algorithms to identify potential unintended targets
- Efficiency prediction: models to estimate editing efficiency
- Repair outcome prediction: Predicting the spectrum of editing outcomes
- Visualization tools: Representing complex genomic targeting information
- Data integration: Combining sgRNA design with functional genomics data
• Structure and components:
- Spacer sequence (20 nucleotides): Complementary to the target DNA sequence
- Scaffold sequence (~80 nucleotides): Forms secondary structures necessary for Cas protein binding
- PAM (Protospacer Adjacent Motif): DNA sequence required for target recognition (not part of sgRNA but essential for targeting)
• Design considerations:
- Target specificity: Minimizing off-target effects through careful sequence selection
- GC content: Optimal range of 40-60% for efficient binding
- Secondary structure: Avoiding self-complementarity that could interfere with target binding
- Position effects: Targeting the beginning of genes or critical functional
- PAM proximity: Selecting targets with appropriate PAM sequences for the Cas variant used
• Bioinformatic tools for sgRNA design:
- CHOPCHOP: Web tool for CRISPR/Cas9 target prediction and off-target evaluation
- CRISPOR: Comprehensive tool for guide selection and off-target prediction
- E-CRISP: Design tool with evaluation of on-target efficiency and off-target effects
- Cas-Designer: Tool for designing guide RNAs for various CRISPR systems
- CRISPRscan: Algorithm for predicting sgRNA efficiency in vivo
• Applications in genome editing:
- Gene knockout: Introducing frameshift mutations through NHEJ repair
- Gene knock-in: Precise sequence insertion via HDR pathway
- Base editing: Creating specific nucleotide changes without double-strand breaks
- Epigenetic modification: Targeting chromatin modifiers to specific genomic loci
- Transcriptional regulation: Activating or repressing gene expression (CRISPRa/CRISPRi)
- Multiplexed editing: Simultaneous modification of multiple genomic targets
• Computational challenges:
- Off-target prediction: Algorithms to identify potential unintended targets
- Efficiency prediction: models to estimate editing efficiency
- Repair outcome prediction: Predicting the spectrum of editing outcomes
- Visualization tools: Representing complex genomic targeting information
- Data integration: Combining sgRNA design with functional genomics data
🔒🧮 SHA-256 Checksum
computer science
SHA-256 (Secure Hash Algorithm 256-bit) is a cryptographic hash function that generates a fixed-size 256-bit (32-byte) hash value, typically rendered as a 64-character hexadecimal string. It belongs to the SHA-2 family of hash functions, designed by the National Security Agency (NSA). SHA-256 is widely used for data integrity verification through checksums, which allow users to verify that a file or message hasn't been altered during transmission or storage. It possesses several critical properties: it's deterministic (the same input always produces the same output), fast to compute, designed to be collision-resistant (extremely difficult to find two different inputs that produce the same hash), and exhibits the avalanche effect (a small change in input drastically changes the output). SHA-256 is extensively used in digital signatures, blockchain technology (particularly Bitcoin), password storage, SSL/TLS certificates, and file verification systems. Unlike encryption, SHA-256 is a one-way function, meaning it's computationally infeasible to reverse the process and derive the original input from the hash value.
🔗 Similarity Network Fusion (SNF)
machine-learningcomputer science
• Definition: A computational method that integrates multiple data types to create a unified patient similarity network, enabling more comprehensive analysis than single-data approaches.
• Algorithm principles:
- Constructs similarity networks for each data type separately
- Iteratively updates each network by fusing information from other networks
- Converges to a single integrated network that captures complementary information across data types
- Uses spectral clustering for patient stratification and subtype identification
• Applications in multi-omics integration:
- Cancer subtyping: Identifying disease subtypes by integrating genomic, transcriptomic, and clinical data
- Biomarker discovery: Finding robust biomarkers across multiple data platforms
- Patient stratification: Grouping patients with similar molecular profiles across different data types
- Drug response prediction: Integrating molecular and pharmacological data to predict treatment outcomes
• Advantages over single-data analysis:
- Increased statistical power through data integration
- Robustness to noise in individual data types
- Ability to capture complementary information across heterogeneous data
- Improved prediction accuracy for clinical outcomes
• Implementation considerations:
- Parameter selection (number of neighbors, fusion iterations)
- Data normalization across different platforms
- Computational efficiency for large datasets
- Visualization of integrated networks
• Algorithm principles:
- Constructs similarity networks for each data type separately
- Iteratively updates each network by fusing information from other networks
- Converges to a single integrated network that captures complementary information across data types
- Uses spectral clustering for patient stratification and subtype identification
• Applications in multi-omics integration:
- Cancer subtyping: Identifying disease subtypes by integrating genomic, transcriptomic, and clinical data
- Biomarker discovery: Finding robust biomarkers across multiple data platforms
- Patient stratification: Grouping patients with similar molecular profiles across different data types
- Drug response prediction: Integrating molecular and pharmacological data to predict treatment outcomes
• Advantages over single-data analysis:
- Increased statistical power through data integration
- Robustness to noise in individual data types
- Ability to capture complementary information across heterogeneous data
- Improved prediction accuracy for clinical outcomes
• Implementation considerations:
- Parameter selection (number of neighbors, fusion iterations)
- Data normalization across different platforms
- Computational efficiency for large datasets
- Visualization of integrated networks
🔓 Small Subgroup Vulnerabilities
computer sciencemath
A cryptographic weakness that can occur in implementations of protocols using discrete logarithm-based cryptography, particularly Diffie-Hellman key exchange. These vulnerabilities arise when an attacker forces computations into a small subgroup of the larger cryptographic group, making it feasible to determine the private key through brute force methods.
Small subgroup attacks exploit improper parameter validation, specifically when implementations fail to verify that received public keys are members of the correct cryptographic group of appropriate order. By sending carefully crafted invalid public values, attackers can extract information about the victim's private key through multiple protocol interactions. Proper implementation requires validation of all public keys and the use of safe primes or prime order subgroups to mitigate these vulnerabilities.
Small subgroup attacks exploit improper parameter validation, specifically when implementations fail to verify that received public keys are members of the correct cryptographic group of appropriate order. By sending carefully crafted invalid public values, attackers can extract information about the victim's private key through multiple protocol interactions. Proper implementation requires validation of all public keys and the use of safe primes or prime order subgroups to mitigate these vulnerabilities.
📊 Softmax
machine-learningmath
A mathematical function that converts a vector of real numbers into a probability distribution, where each output value is between 0 and 1 and all outputs sum to 1. Softmax is commonly used as the final activation function in multi-class classification problems, transforming raw model outputs (logits) into interpretable probabilities for each class.
Mathematically defined as: for all j, where x is the input vector. The exponential function ensures all outputs are positive, while the normalization by the sum creates a valid probability distribution. Softmax amplifies the differences between values - larger inputs receive disproportionately higher probabilities, making it useful for confident predictions.
Softmax is essential in for tasks like image classification (determining which of several objects appears in an image), natural language processing (predicting the next word from a vocabulary), and any scenario requiring probabilistic outputs across multiple mutually exclusive categories. It's often paired with cross-entropy loss during training to optimize classification performance.
Mathematically defined as: for all j, where x is the input vector. The exponential function ensures all outputs are positive, while the normalization by the sum creates a valid probability distribution. Softmax amplifies the differences between values - larger inputs receive disproportionately higher probabilities, making it useful for confident predictions.
Softmax is essential in for tasks like image classification (determining which of several objects appears in an image), natural language processing (predicting the next word from a vocabulary), and any scenario requiring probabilistic outputs across multiple mutually exclusive categories. It's often paired with cross-entropy loss during training to optimize classification performance.
🧬 Somatic Mutations
biology
• Definition: Alterations in DNA that occur after conception in somatic (non-germline) cells of the body and are not inherited from parents or passed to offspring. These mutations accumulate throughout an individual's lifetime in various tissues due to environmental exposures, replication errors, or processes.
• Comparison with germline mutations:
- Somatic mutations: Present only in specific cells/tissues, not in every cell of the body, not heritable
- Germline mutations: Present in germ cells (sperm or eggs), found in every cell of the body, can be passed to offspring
- Distribution: Somatic mutations show tissue-specific patterns and increase with age, while germline variants are uniformly distributed across tissues
• Types of somatic mutations:
- Driver mutations: Confer selective growth advantage to cells, contribute directly to cancer development by affecting oncogenes or tumor suppressor genes
- Passenger mutations: Do not provide growth advantage, occur coincidentally in cells that already have driver mutations
- Based on DNA change: Single nucleotide variants (SNVs), insertions/deletions (indels), copy number alterations (CNAs), structural variants (SVs), and chromosomal rearrangements
• Mutational signatures:
- Characteristic patterns of mutations caused by specific mutational processes
- Examples: UV light exposure (C>T transitions), tobacco smoke (G>T transversions), APOBEC enzyme activity, defective DNA repair mechanisms
- Provide insights into cancer etiology and progression mechanisms
• Detection methods:
- Next-generation sequencing (NGS): Whole-genome, whole-exome, or targeted panel sequencing
- Tumor-normal comparison: Sequencing both tumor and matched normal tissue to identify somatic-specific variants
- Deep sequencing: Required to detect low-frequency mutations in heterogeneous samples
- Single-cell sequencing: For analyzing intratumoral heterogeneity and clonal evolution
- Liquid biopsy: Detection of circulating tumor DNA (ctDNA) in blood for non-invasive monitoring
• Clinical significance:
- Cancer diagnosis: Identifying driver mutations that define cancer subtypes
- Prognostic biomarkers: Mutations associated with disease progression or survival outcomes
- Predictive biomarkers: Mutations that predict response to specific therapies
- Therapeutic targets: Basis for precision medicine approaches in cancer treatment
- Monitoring: Tracking treatment response and disease recurrence through serial sampling
• Applications in precision oncology:
- Targeted therapies: Drugs designed to target specific mutations (e.g., EGFR inhibitors for EGFR mutations)
- Immunotherapy selection: Tumor mutational burden (TMB) as a biomarker for immunotherapy response
- Pan-cancer approaches: Targeting mutations across multiple cancer types regardless of tissue of origin
- Resistance mechanisms: Identifying secondary mutations that confer treatment resistance
- Minimal residual disease detection: Using somatic mutations as personalized biomarkers
• Computational challenges:
- Distinguishing driver from passenger mutations
- Accounting for tumor heterogeneity and normal cell contamination
- Identifying mutations in low-frequency subclones
- Integrating multiple data types for comprehensive mutational profiling
- Interpreting variants of unknown significance
• Comparison with germline mutations:
- Somatic mutations: Present only in specific cells/tissues, not in every cell of the body, not heritable
- Germline mutations: Present in germ cells (sperm or eggs), found in every cell of the body, can be passed to offspring
- Distribution: Somatic mutations show tissue-specific patterns and increase with age, while germline variants are uniformly distributed across tissues
• Types of somatic mutations:
- Driver mutations: Confer selective growth advantage to cells, contribute directly to cancer development by affecting oncogenes or tumor suppressor genes
- Passenger mutations: Do not provide growth advantage, occur coincidentally in cells that already have driver mutations
- Based on DNA change: Single nucleotide variants (SNVs), insertions/deletions (indels), copy number alterations (CNAs), structural variants (SVs), and chromosomal rearrangements
• Mutational signatures:
- Characteristic patterns of mutations caused by specific mutational processes
- Examples: UV light exposure (C>T transitions), tobacco smoke (G>T transversions), APOBEC enzyme activity, defective DNA repair mechanisms
- Provide insights into cancer etiology and progression mechanisms
• Detection methods:
- Next-generation sequencing (NGS): Whole-genome, whole-exome, or targeted panel sequencing
- Tumor-normal comparison: Sequencing both tumor and matched normal tissue to identify somatic-specific variants
- Deep sequencing: Required to detect low-frequency mutations in heterogeneous samples
- Single-cell sequencing: For analyzing intratumoral heterogeneity and clonal evolution
- Liquid biopsy: Detection of circulating tumor DNA (ctDNA) in blood for non-invasive monitoring
• Clinical significance:
- Cancer diagnosis: Identifying driver mutations that define cancer subtypes
- Prognostic biomarkers: Mutations associated with disease progression or survival outcomes
- Predictive biomarkers: Mutations that predict response to specific therapies
- Therapeutic targets: Basis for precision medicine approaches in cancer treatment
- Monitoring: Tracking treatment response and disease recurrence through serial sampling
• Applications in precision oncology:
- Targeted therapies: Drugs designed to target specific mutations (e.g., EGFR inhibitors for EGFR mutations)
- Immunotherapy selection: Tumor mutational burden (TMB) as a biomarker for immunotherapy response
- Pan-cancer approaches: Targeting mutations across multiple cancer types regardless of tissue of origin
- Resistance mechanisms: Identifying secondary mutations that confer treatment resistance
- Minimal residual disease detection: Using somatic mutations as personalized biomarkers
• Computational challenges:
- Distinguishing driver from passenger mutations
- Accounting for tumor heterogeneity and normal cell contamination
- Identifying mutations in low-frequency subclones
- Integrating multiple data types for comprehensive mutational profiling
- Interpreting variants of unknown significance
🔐💻 SSH (Secure Shell)
computer science
SSH is a cryptographic network protocol that provides secure communication over an unsecured network by establishing an encrypted connection between two computers. Developed as a replacement for insecure protocols like Telnet and rsh, SSH uses public-key cryptography for authentication and symmetric encryption for confidentiality and integrity of data transmission. The protocol operates primarily on TCP 22 and supports multiple authentication methods, including password-based authentication, public key authentication, host-based authentication, and keyboard-interactive authentication. SSH enables several secure network services: remote command-line login, remote command execution, secure file transfer (via SFTP or SCP), forwarding (tunneling), and X11 forwarding for running graphical applications remotely. It's widely used by system administrators for secure remote server management, by developers for secure access to version control systems, and as a fundamental component in automated deployment pipelines. SSH's architecture consists of three major components: the transport layer protocol (providing encryption and server authentication), the user authentication protocol, and the connection protocol (multiplexing the encrypted tunnel into multiple logical channels). Modern implementations like OpenSSH have become the de facto standard for secure remote access across Unix-like operating systems, Windows, and network devices.
🔄 Stereoisomers
biology
• Definition: Isomers that have the same molecular formula and sequence of bonded atoms but differ in the three-dimensional orientation of their atoms in space.
• Major types:
- Enantiomers: Mirror-image stereoisomers that are non-superimposable
- Diastereomers: Non-mirror-image stereoisomers
- Conformational isomers: Stereoisomers that can interconvert by rotation around single bonds
- Geometric isomers (cis-trans): Differ in the arrangement of groups around a double bond or ring
• Biological significance:
- Molecular recognition: Biological systems can distinguish between stereoisomers
- Pharmacology: Different stereoisomers can have different pharmacological properties
- Metabolism: Enzymes often process only specific stereoisomers
- Structural biology: Protein folding and nucleic acid structure depend on specific stereochemistry
• Analytical methods:
- X-ray crystallography: Determining absolute configuration
• Major types:
- Enantiomers: Mirror-image stereoisomers that are non-superimposable
- Diastereomers: Non-mirror-image stereoisomers
- Conformational isomers: Stereoisomers that can interconvert by rotation around single bonds
- Geometric isomers (cis-trans): Differ in the arrangement of groups around a double bond or ring
• Biological significance:
- Molecular recognition: Biological systems can distinguish between stereoisomers
- Pharmacology: Different stereoisomers can have different pharmacological properties
- Metabolism: Enzymes often process only specific stereoisomers
- Structural biology: Protein folding and nucleic acid structure depend on specific stereochemistry
• Analytical methods:
- X-ray crystallography: Determining absolute configuration
🎲 Stochastic
math
In simple terms, "stochastic" refers to processes or systems that are random or inherently unpredictable, but they follow some statistical patterns or probabilities. Stochastic systems don't have a single, fixed outcome; instead, their behavior or outcomes are governed by chance and probability distributions.
The term is widely used in fields like mathematics, biology, finance, and physics, where randomness and uncertainty are present.
The term is widely used in fields like mathematics, biology, finance, and physics, where randomness and uncertainty are present.
🌐 Subdomain
networkingweb-development
A prefix added to a name that creates a separate section or service within the main . Subdomains allow organizations to organize different services, departments, or geographical regions under their primary .
Subdomains appear before the main name, separated by a dot. For example, in 'blog.example.com', 'blog' is the subdomain of 'example.com'.
Common Subdomain Examples:
- www: Traditional web server (www.example.com)
- mail: Email services (mail.example.com)
- blog: Blog or news section (blog.example.com)
- api: API endpoints (api.example.com)
- cdn: Content delivery network (cdn.example.com)
- staging: Development/testing environment (staging.example.com)
Subdomains can have their own DNS records, SSL certificates, and can point to different servers or IP addresses than the main . They're useful for organizing content, load balancing, and creating logical separations within a website's architecture.
Subdomains appear before the main name, separated by a dot. For example, in 'blog.example.com', 'blog' is the subdomain of 'example.com'.
Common Subdomain Examples:
- www: Traditional web server (www.example.com)
- mail: Email services (mail.example.com)
- blog: Blog or news section (blog.example.com)
- api: API endpoints (api.example.com)
- cdn: Content delivery network (cdn.example.com)
- staging: Development/testing environment (staging.example.com)
Subdomains can have their own DNS records, SSL certificates, and can point to different servers or IP addresses than the main . They're useful for organizing content, load balancing, and creating logical separations within a website's architecture.
👨🏫 Supervised Learning
machine-learning
A learning approach where the AI system is trained on labeled data (examples with known correct answers). It's like learning with a teacher who provides the correct answers. This is the most common form of , used in applications like spam detection and image classification.
🧪 Svedberg Sedimentation Coefficient
biologylab-techniques
• Definition: A unit of measurement (symbol S) that characterizes the sedimentation rate of particles under centrifugal force, defined as the ratio of a particle's sedimentation velocity to the applied acceleration. One Svedberg unit equals exactly 10⁻¹³ seconds (100 femtoseconds).
• Key principles:
- Named after Swedish chemist Theodor Svedberg, who won the 1926 Nobel Prize for his work on colloids and invention of the ultracentrifuge
- Represents how quickly a particle settles in solution during centrifugation
- Determined by a particle's mass, density, and shape
- Larger, heavier particles generally have higher S values
- Svedberg values are not additive; when two particles bind together, their combined S value is not the sum of their individual values
• Mathematical basis:
- Described by the Svedberg equation: s = m(1-νρ)/f
- Where m is the particle mass, ν is the partial specific volume, ρ is the solvent density, and f is the frictional coefficient
- For a spherical particle, the frictional coefficient is related to its radius by Stokes' law
- A particle with a sedimentation coefficient of 26S will travel at 26 micrometers per second under an acceleration of 1 million gravities
• Applications in bioinformatics and molecular biology:
- Classification of cellular components like ribosomes (e.g., 70S prokaryotic ribosomes, 80S eukaryotic ribosomes)
- Characterization of ribosomal subunits (e.g., 30S and 50S in prokaryotes, 40S and 60S in eukaryotes)
- Identification of protein complexes and determination of their stoichiometry
- Analysis of macromolecular interactions and binding affinities
- Determination of size distributions in heterogeneous samples
• Analytical techniques:
- Analytical ultracentrifugation (AUC) is the primary method for measuring sedimentation coefficients
- Sedimentation velocity experiments track boundary movement over time
- Sedimentation equilibrium experiments analyze concentration gradients at equilibrium
- Modern analysis uses computational software to fit data to the Lamm equation
- Detection methods include absorbance, interference, and fluorescence optics
• Significance in structural biology:
- Provides hydrodynamic information complementary to other structural techniques
- Helps determine molecular weight, shape, and conformational changes
- Enables study of macromolecules in their native state without interaction with matrices or surfaces
- Applicable to a wide range of molecular weights (from hundreds to millions of Daltons)
- Allows analysis of complex biological samples including cell lysates and bodily fluids
• Key principles:
- Named after Swedish chemist Theodor Svedberg, who won the 1926 Nobel Prize for his work on colloids and invention of the ultracentrifuge
- Represents how quickly a particle settles in solution during centrifugation
- Determined by a particle's mass, density, and shape
- Larger, heavier particles generally have higher S values
- Svedberg values are not additive; when two particles bind together, their combined S value is not the sum of their individual values
• Mathematical basis:
- Described by the Svedberg equation: s = m(1-νρ)/f
- Where m is the particle mass, ν is the partial specific volume, ρ is the solvent density, and f is the frictional coefficient
- For a spherical particle, the frictional coefficient is related to its radius by Stokes' law
- A particle with a sedimentation coefficient of 26S will travel at 26 micrometers per second under an acceleration of 1 million gravities
• Applications in bioinformatics and molecular biology:
- Classification of cellular components like ribosomes (e.g., 70S prokaryotic ribosomes, 80S eukaryotic ribosomes)
- Characterization of ribosomal subunits (e.g., 30S and 50S in prokaryotes, 40S and 60S in eukaryotes)
- Identification of protein complexes and determination of their stoichiometry
- Analysis of macromolecular interactions and binding affinities
- Determination of size distributions in heterogeneous samples
• Analytical techniques:
- Analytical ultracentrifugation (AUC) is the primary method for measuring sedimentation coefficients
- Sedimentation velocity experiments track boundary movement over time
- Sedimentation equilibrium experiments analyze concentration gradients at equilibrium
- Modern analysis uses computational software to fit data to the Lamm equation
- Detection methods include absorbance, interference, and fluorescence optics
• Significance in structural biology:
- Provides hydrodynamic information complementary to other structural techniques
- Helps determine molecular weight, shape, and conformational changes
- Enables study of macromolecules in their native state without interaction with matrices or surfaces
- Applicable to a wide range of molecular weights (from hundreds to millions of Daltons)
- Allows analysis of complex biological samples including cell lysates and bodily fluids
🎯 Targeted Protein Degradation
biology
• Definition: A therapeutic strategy that harnesses the cell's protein degradation machinery to selectively eliminate disease-causing proteins, particularly those considered "undruggable" by traditional approaches.
• Major degradation pathways utilized:
- Ubiquitin-proteasome system (UPS): Primary pathway for degradation of intracellular proteins
- Lysosomal pathway: Alternative route for degradation of membrane proteins and aggregates
- Autophagy: Bulk degradation of cellular components including organelles and protein aggregates
• Key technologies:
- PROteolysis TArgeting Chimeras (PROTACs): Bifunctional molecules linking target proteins to E3 ligases
- Molecular glues: Monovalent molecules that enhance protein-protein interactions with E3 ligases
- Lysosome-targeting chimeras (LYTACs): Direct proteins to lysosomal degradation
- Autophagy-targeting chimeras (AUTACs): Target proteins for autophagic degradation
- dTAGs: Degradation tags for rapid protein elimination
• Advantages over traditional inhibitors:
- Catalytic mode of action: One degrader molecule can process multiple target proteins
- Event-driven rather than occupancy-driven: Prolonged effect after drug clearance
- Ability to target non-enzymatic protein functions and scaffolding proteins
- Potential to overcome resistance mechanisms
- Lower required drug concentrations for efficacy
• Challenges and limitations:
- Tissue distribution and intracellular penetration
- Identification of appropriate E3 ligases for specific tissues
- Potential for off-target effects
- Complex pharmacokinetic and pharmacodynamic relationships
• Clinical applications:
- Oncology: Degradation of oncoproteins and transcription factors
- Neurodegenerative diseases: Clearance of protein aggregates
- Inflammatory disorders: Degradation of inflammatory mediators
- Infectious diseases: Elimination of viral proteins
• Major degradation pathways utilized:
- Ubiquitin-proteasome system (UPS): Primary pathway for degradation of intracellular proteins
- Lysosomal pathway: Alternative route for degradation of membrane proteins and aggregates
- Autophagy: Bulk degradation of cellular components including organelles and protein aggregates
• Key technologies:
- PROteolysis TArgeting Chimeras (PROTACs): Bifunctional molecules linking target proteins to E3 ligases
- Molecular glues: Monovalent molecules that enhance protein-protein interactions with E3 ligases
- Lysosome-targeting chimeras (LYTACs): Direct proteins to lysosomal degradation
- Autophagy-targeting chimeras (AUTACs): Target proteins for autophagic degradation
- dTAGs: Degradation tags for rapid protein elimination
• Advantages over traditional inhibitors:
- Catalytic mode of action: One degrader molecule can process multiple target proteins
- Event-driven rather than occupancy-driven: Prolonged effect after drug clearance
- Ability to target non-enzymatic protein functions and scaffolding proteins
- Potential to overcome resistance mechanisms
- Lower required drug concentrations for efficacy
• Challenges and limitations:
- Tissue distribution and intracellular penetration
- Identification of appropriate E3 ligases for specific tissues
- Potential for off-target effects
- Complex pharmacokinetic and pharmacodynamic relationships
• Clinical applications:
- Oncology: Degradation of oncoproteins and transcription factors
- Neurodegenerative diseases: Clearance of protein aggregates
- Inflammatory disorders: Degradation of inflammatory mediators
- Infectious diseases: Elimination of viral proteins
🤝📬 TCP (Transmission Control Protocol)
computer science
TCP is a connection-oriented protocol that operates at the transport layer of the Internet Protocol Suite. It provides reliable, ordered, and error-checked delivery of data between applications running on hosts communicating over an IP network. TCP establishes a full-duplex communication channel between sender and receiver through a process called a three-way handshake (SYN, SYN-ACK, ACK) before any data exchange begins. It implements flow control through a sliding window mechanism, congestion control to prevent network overload, and retransmission of lost packets to ensure data integrity. TCP segments data into packets, assigns sequence numbers for proper ordering, and requires acknowledgment of received packets. Due to these reliability features, TCP is used for applications where accurate data delivery is critical, such as web browsing (HTTP/HTTPS), email (SMTP), file transfers (FTP), and remote administration (SSH). The trade-off for this reliability is increased latency and overhead compared to connectionless protocols like UDP.
📊 Tensor Decomposition
machine-learning
A mathematical approach for breaking down multi-dimensional arrays (tensors) into simpler components, similar to how matrix factorization works for two-dimensional data. Tensor decompositions reveal underlying patterns and structures in complex multi-way data, making them powerful tools for dimensionality reduction, feature extraction, and latent factor discovery in .
Tensor methods are particularly valuable for analyzing data with multiple aspects or modes (such as users-items-time in recommendation systems or subjects-features-time in neuroimaging), where traditional matrix methods would lose important multi-way relationships.
Tensor methods are particularly valuable for analyzing data with multiple aspects or modes (such as users-items-time in recommendation systems or subjects-features-time in neuroimaging), where traditional matrix methods would lose important multi-way relationships.
🔢 Tensor Processing Unit (TPU)
machine-learning
A specialized hardware accelerator developed by Google specifically for . TPUs are application-specific integrated circuits (ASICs) designed to accelerate tensor operations, which are fundamental to models. Unlike CPUs and GPUs, TPUs are optimized for matrix operations with high computational throughput and lower precision, making them significantly faster and more energy-efficient for AI workloads.
TPUs play a crucial role in training and running Google's Gemini models, providing the massive computational power needed to process the enormous datasets required for large language models. Their architecture enables parallel processing at scale, reducing training time from weeks to days or hours, and allowing for more efficient inference when deploying these models in production environments.
TPUs play a crucial role in training and running Google's Gemini models, providing the massive computational power needed to process the enormous datasets required for large language models. Their architecture enables parallel processing at scale, reducing training time from weeks to days or hours, and allowing for more efficient inference when deploying these models in production environments.
🔒 TLS (Transport Layer Security)
computer sciencesecurity
A cryptographic protocol that provides secure communication over a computer network. TLS is the successor to SSL (Secure Sockets Layer) and is widely used to secure internet communications, including web browsing, email, instant messaging, and voice over IP (VoIP).
TLS works by establishing an encrypted connection between a client and server through a process called the TLS handshake. During this handshake, the client and server:
1. Negotiate protocol version and cipher suites
2. Authenticate the server (and optionally the client) using digital certificates
3. Exchange cryptographic keys to establish a secure session
4. Begin encrypted communication using symmetric encryption
HTTPS = HTTP + TLS
When TLS is applied to HTTP (Hypertext Transfer Protocol), it creates HTTPS (HTTP Secure). This combination ensures that all data transmitted between a web browser and web server is encrypted and authenticated. The 'S' in HTTPS specifically indicates that the HTTP connection is secured using TLS/SSL encryption.
HTTPS operates on 443 (compared to HTTP's 80) and provides three key security benefits:
- Confidentiality: Data is encrypted and cannot be read by eavesdroppers
- Integrity: Data cannot be modified without detection
- Authentication: Verifies the identity of the communicating parties
TLS is essential for protecting sensitive information like passwords, credit card numbers, and personal data transmitted over the internet. Modern web browsers display security indicators (like a padlock icon) to show when a connection is secured with TLS.
TLS works by establishing an encrypted connection between a client and server through a process called the TLS handshake. During this handshake, the client and server:
1. Negotiate protocol version and cipher suites
2. Authenticate the server (and optionally the client) using digital certificates
3. Exchange cryptographic keys to establish a secure session
4. Begin encrypted communication using symmetric encryption
HTTPS = HTTP + TLS
When TLS is applied to HTTP (Hypertext Transfer Protocol), it creates HTTPS (HTTP Secure). This combination ensures that all data transmitted between a web browser and web server is encrypted and authenticated. The 'S' in HTTPS specifically indicates that the HTTP connection is secured using TLS/SSL encryption.
HTTPS operates on 443 (compared to HTTP's 80) and provides three key security benefits:
- Confidentiality: Data is encrypted and cannot be read by eavesdroppers
- Integrity: Data cannot be modified without detection
- Authentication: Verifies the identity of the communicating parties
TLS is essential for protecting sensitive information like passwords, credit card numbers, and personal data transmitted over the internet. Modern web browsers display security indicators (like a padlock icon) to show when a connection is secured with TLS.
✂️ Tokenization
machine-learning
The process of breaking text into smaller units (tokens) that can be processed by AI models. These tokens might be words, subwords, or characters. Modern systems often use subword tokenization to handle unknown words and reduce vocabulary size while maintaining meaning.
🧬 Transcription Factor (TF) Networks
biology
• Purpose: Regulatory systems composed of transcription factors that control gene expression by binding to specific DNA sequences.
• Structure: Complex networks where TFs interact with DNA and with each other to form regulatory circuits that determine when and where genes are expressed.
• Key components:
- Transcription factors: Proteins that bind to specific DNA sequences to control gene transcription
- Binding motifs: Short DNA sequences recognized by specific TFs
- Network motifs: Recurring patterns in regulatory networks (e.g., feed-forward loops, feedback loops)
- Hierarchical organization: Master regulators controlling other TFs, creating regulatory cascades
• Biological significance: TF networks orchestrate critical processes including cell differentiation, development, and response to environmental signals.
• Applications in bioinformatics:
- Network inference: Computational methods to reconstruct TF networks from genomic data
- Motif discovery: Identifying DNA sequences bound by TFs
- Network analysis: Understanding regulatory relationships and predicting cellular responses
- Disease research: Identifying dysregulated TF networks in conditions like cancer and developmental disorders
• Structure: Complex networks where TFs interact with DNA and with each other to form regulatory circuits that determine when and where genes are expressed.
• Key components:
- Transcription factors: Proteins that bind to specific DNA sequences to control gene transcription
- Binding motifs: Short DNA sequences recognized by specific TFs
- Network motifs: Recurring patterns in regulatory networks (e.g., feed-forward loops, feedback loops)
- Hierarchical organization: Master regulators controlling other TFs, creating regulatory cascades
• Biological significance: TF networks orchestrate critical processes including cell differentiation, development, and response to environmental signals.
• Applications in bioinformatics:
- Network inference: Computational methods to reconstruct TF networks from genomic data
- Motif discovery: Identifying DNA sequences bound by TFs
- Network analysis: Understanding regulatory relationships and predicting cellular responses
- Disease research: Identifying dysregulated TF networks in conditions like cancer and developmental disorders
🔄 Transformer
machine-learning
An influential architecture that revolutionized natural language processing. It uses self- to process sequential data, enabling better understanding of context in language. Transformers power most modern language models like GPT and BERT.
🧩 Tucker Decomposition
mathmachine-learning
A higher-order extension of principal component analysis (PCA) that decomposes a tensor into a core tensor multiplied by a matrix along each mode. Tucker decomposition provides a more flexible representation than other tensor methods, allowing different ranks for different dimensions.
In , Tucker decomposition excels at subspace learning and dimensionality reduction for multi-way data, enabling applications like multi-aspect data mining, anomaly detection in network traffic, and feature extraction from multi-modal signals.
In , Tucker decomposition excels at subspace learning and dimensionality reduction for multi-way data, enabling applications like multi-aspect data mining, anomaly detection in network traffic, and feature extraction from multi-modal signals.
🔄 Ubiquitin Proteasome System (UPS)
biology
• Definition: A major intracellular protein degradation pathway that selectively tags proteins with ubiquitin molecules for subsequent degradation by the 26S proteasome complex, playing a crucial role in protein homeostasis and cellular quality control.
• Key components:
- Ubiquitin: A 76-amino acid protein that serves as the tag for protein degradation
- E1 (ubiquitin-activating enzymes): Activate ubiquitin in an ATP-dependent manner
- E2 (ubiquitin-conjugating enzymes): Transfer activated ubiquitin from E1
- E3 (ubiquitin ligases): Recognize specific substrates and facilitate ubiquitin transfer
- Proteasome: A large multi-subunit complex that degrades ubiquitinated proteins
- Deubiquitinating enzymes (DUBs): Remove ubiquitin tags, providing regulation
• Ubiquitination process:
- Activation: E1 activates ubiquitin in an ATP-dependent reaction
- Conjugation: E2 accepts the activated ubiquitin from E1
- Ligation: E3 catalyzes the transfer of ubiquitin to lysine residues on target proteins
- Polyubiquitination: Multiple ubiquitin molecules form chains (K48-linked chains target for degradation)
• Proteasomal degradation:
- Recognition: Polyubiquitinated proteins are recognized by the 19S regulatory particle
- Unfolding: Proteins are unfolded and deubiquitinated
- Degradation: Unfolded proteins enter the 20S core particle and are cleaved into peptides
- Recycling: Amino acids are recycled for new protein synthesis
• Biological significance:
- Protein quality control: Elimination of misfolded or damaged proteins
- Regulation of protein abundance: Control of cellular levels of regulatory proteins
- Cell cycle control: Degradation of cyclins and cell cycle inhibitors
- Signal transduction: Regulation of signaling pathway components
- Immune response: Antigen processing for MHC class I presentation
• Clinical relevance:
- Cancer: Proteasome inhibitors (e.g., bortezomib) approved for multiple myeloma
- Neurodegenerative diseases: Accumulation of protein aggregates due to UPS dysfunction
- Inflammatory disorders: UPS involvement in NF-κB pathway regulation
- Emerging target for drug development: approaches
• Key components:
- Ubiquitin: A 76-amino acid protein that serves as the tag for protein degradation
- E1 (ubiquitin-activating enzymes): Activate ubiquitin in an ATP-dependent manner
- E2 (ubiquitin-conjugating enzymes): Transfer activated ubiquitin from E1
- E3 (ubiquitin ligases): Recognize specific substrates and facilitate ubiquitin transfer
- Proteasome: A large multi-subunit complex that degrades ubiquitinated proteins
- Deubiquitinating enzymes (DUBs): Remove ubiquitin tags, providing regulation
• Ubiquitination process:
- Activation: E1 activates ubiquitin in an ATP-dependent reaction
- Conjugation: E2 accepts the activated ubiquitin from E1
- Ligation: E3 catalyzes the transfer of ubiquitin to lysine residues on target proteins
- Polyubiquitination: Multiple ubiquitin molecules form chains (K48-linked chains target for degradation)
• Proteasomal degradation:
- Recognition: Polyubiquitinated proteins are recognized by the 19S regulatory particle
- Unfolding: Proteins are unfolded and deubiquitinated
- Degradation: Unfolded proteins enter the 20S core particle and are cleaved into peptides
- Recycling: Amino acids are recycled for new protein synthesis
• Biological significance:
- Protein quality control: Elimination of misfolded or damaged proteins
- Regulation of protein abundance: Control of cellular levels of regulatory proteins
- Cell cycle control: Degradation of cyclins and cell cycle inhibitors
- Signal transduction: Regulation of signaling pathway components
- Immune response: Antigen processing for MHC class I presentation
• Clinical relevance:
- Cancer: Proteasome inhibitors (e.g., bortezomib) approved for multiple myeloma
- Neurodegenerative diseases: Accumulation of protein aggregates due to UPS dysfunction
- Inflammatory disorders: UPS involvement in NF-κB pathway regulation
- Emerging target for drug development: approaches
🚀📨 UDP (User Datagram Protocol)
computer science
UDP is a connectionless transport layer protocol in the Internet Protocol Suite that operates with minimal overhead and no guaranteed delivery. Unlike TCP, UDP does not establish a connection before sending data, does not provide reliability mechanisms like acknowledgments or retransmissions, and does not guarantee that packets (called datagrams) arrive in the same order they were sent. UDP simply transmits datagrams without verifying receipt, making it significantly faster and more efficient than TCP but less reliable. The protocol includes only basic error checking through checksums but does not attempt to recover from errors. UDP is ideal for applications where speed is more important than perfect reliability, such as real-time applications that can tolerate some data loss: video streaming, voice over IP (VoIP), online gaming, DNS lookups, and IoT communications. Its low overhead makes it particularly suitable for applications that send small amounts of data or require minimal latency.
🔍 Unsupervised Learning
machine-learning
A learning method where the AI system finds patterns in unlabeled data without explicit guidance. It's like discovering categories or relationships naturally. Important for data clustering, anomaly detection, and understanding hidden patterns in data.
🔗 URL (Uniform Resource Locator)
networkingweb-development
A complete web address that specifies the exact location of a resource on the internet. URLs provide a standardized way to access web pages, files, images, and other resources across the web.
URL Structure:
`protocol://..tld:/path?query=value#anchor`
Components Breakdown:
- Protocol/Scheme: How to access the resource (http, https, ftp)
- ****: Optional prefix to the (www, api, blog)
- ****: The main website address (example.com)
- ****: Optional network number (default: 80 for HTTP, 443 for HTTPS)
- Path: Specific page or resource location (/about/team)
- Query Parameters: Additional data sent to the server (?search=term&page=2)
- Anchor/Fragment: Specific section within a page (#section1)
Example URL:
`https://www.example.com:443/products/shoes?color=blue&size=10#reviews`
URLs are essential for web navigation, linking, bookmarking, and API endpoints. They must follow specific encoding rules to handle special characters and spaces.
URL Structure:
`protocol://..tld:/path?query=value#anchor`
Components Breakdown:
- Protocol/Scheme: How to access the resource (http, https, ftp)
- ****: Optional prefix to the (www, api, blog)
- ****: The main website address (example.com)
- ****: Optional network number (default: 80 for HTTP, 443 for HTTPS)
- Path: Specific page or resource location (/about/team)
- Query Parameters: Additional data sent to the server (?search=term&page=2)
- Anchor/Fragment: Specific section within a page (#section1)
Example URL:
`https://www.example.com:443/products/shoes?color=blue&size=10#reviews`
URLs are essential for web navigation, linking, bookmarking, and API endpoints. They must follow specific encoding rules to handle special characters and spaces.
🗜️ Variational Autoencoder (VAE)
machine-learning
A generative model that learns to compress data into a compact representation and then reconstruct it. Unlike regular autoencoders, VAEs learn smooth, continuous representations that allow meaningful data generation and manipulation. They're vital for tasks like image generation and data compression.
🗄️ Vector Database
machine-learning
A specialized database designed to store and efficiently search through high-dimensional vectors (). These databases enable rapid similarity search and are crucial for modern AI applications like semantic search, recommendation systems, and image retrieval.
🔌💻 Verilog
biology
Verilog is a hardware description language (HDL) used primarily for modeling, simulating, and synthesizing digital circuits and systems. It allows engineers to describe electronic systems at various levels of abstraction, from high-level behavioral descriptions to detailed gate-level implementations. Verilog is widely used in the semiconductor industry for designing application-specific integrated circuits (ASICs), field-programmable gate arrays (FPGAs), and other digital hardware. It supports both concurrent and sequential execution models, reflecting the parallel nature of hardware operations. Along with VHDL, Verilog is one of the two major HDLs used in electronic design automation (EDA) workflows for digital circuit design, verification, and implementation.
🧫🔬 Western Blot/Immunoblot
lab-techniquesbiology
Definition
Western blotting (immunoblotting) is a powerful analytical technique used in molecular biology and proteomics to detect, identify, and semi-quantify specific proteins within a complex mixture of proteins extracted from cells or tissues 1. The technique derives its name from its position in the "blotting" family, following Southern blotting (for DNA) and Northern blotting (for RNA) 1.
Principle
Western blotting uses three key elements to accomplish protein detection 2:
1. Separation by size: Proteins are separated based on molecular weight through gel electrophoresis
2. Transfer to solid support: Separated proteins are transferred to a protein-binding membrane
3. Immunodetection: Target proteins are marked using specific primary and secondary antibodies for visualization
Protocol
The Western blot procedure typically involves the following steps 2 4:
1. Sample preparation: Proteins are extracted from cells or tissues using mechanical disruption, chemical extraction, or enzymatic methods
2. Gel electrophoresis: Proteins are separated based on molecular weight using polyacrylamide gel electrophoresis (PAGE), typically with sodium dodecyl sulfate (SDS-PAGE)
3. Protein transfer: Separated proteins are transferred from the gel to a membrane (nitrocellulose or PVDF) using electrophoresis (electroblotting)
4. Blocking: The membrane is blocked with a protein solution (often non-fat dry milk or BSA) to prevent non-specific antibody binding
5. Primary antibody incubation: The membrane is incubated with a primary antibody specific to the target protein
6. Washing: Unbound primary antibodies are washed away
7. Secondary antibody incubation: The membrane is incubated with a labeled secondary antibody that binds to the primary antibody
8. Detection: The protein-antibody complex is visualized using various detection methods
Detection Methods
Several detection methods can be used in Western blotting 1 4:
1. Chromogenic detection: Uses enzyme-conjugated secondary antibodies (like HRP or AP) that produce a colored precipitate when exposed to a substrate
2. Chemiluminescent detection: Uses enzyme-conjugated antibodies that produce light when exposed to a substrate, captured on film or by digital imaging systems
3. Fluorescent detection: Uses fluorophore-conjugated antibodies that emit light when excited at specific wavelengths
4. Radioactive detection: Historically used radioactive probes and autoradiography film (less common today)
Applications
Western blotting has numerous applications in research and clinical settings 5 3:
1. Protein identification and characterization: Detecting specific proteins in complex mixtures
2. Post-translational modifications: Identifying modifications like phosphorylation, glycosylation, and ubiquitination
3. Protein expression analysis: Measuring relative protein levels in different samples
4. Clinical diagnostics: Confirming diseases like HIV through antibody detection
5. Drug development: Evaluating protein targets and drug effects
6. Epitope mapping: Identifying antibody binding sites on proteins
7. Anti-doping testing: Detecting prohibited substances in sports
Advantages
1. Requires only small amounts of reagents 5
2. High specificity due to antibody-antigen interactions 4
3. Provides information about protein molecular weight 1
4. Same protein transfer can be used for multiple analyses 5
5. Can detect proteins at picogram levels with optimized protocols
Limitations
1. Requires specific antibodies for target proteins 5
2. Potential for antibody cross-reactivity and off-target effects 5
3. Time-consuming procedure (typically 1-2 days) 1
4. Semi-quantitative rather than fully quantitative 1
5. Relatively costly due to antibody expenses and detection reagents 5
Western blotting (immunoblotting) is a powerful analytical technique used in molecular biology and proteomics to detect, identify, and semi-quantify specific proteins within a complex mixture of proteins extracted from cells or tissues 1. The technique derives its name from its position in the "blotting" family, following Southern blotting (for DNA) and Northern blotting (for RNA) 1.
Principle
Western blotting uses three key elements to accomplish protein detection 2:
1. Separation by size: Proteins are separated based on molecular weight through gel electrophoresis
2. Transfer to solid support: Separated proteins are transferred to a protein-binding membrane
3. Immunodetection: Target proteins are marked using specific primary and secondary antibodies for visualization
Protocol
The Western blot procedure typically involves the following steps 2 4:
1. Sample preparation: Proteins are extracted from cells or tissues using mechanical disruption, chemical extraction, or enzymatic methods
2. Gel electrophoresis: Proteins are separated based on molecular weight using polyacrylamide gel electrophoresis (PAGE), typically with sodium dodecyl sulfate (SDS-PAGE)
3. Protein transfer: Separated proteins are transferred from the gel to a membrane (nitrocellulose or PVDF) using electrophoresis (electroblotting)
4. Blocking: The membrane is blocked with a protein solution (often non-fat dry milk or BSA) to prevent non-specific antibody binding
5. Primary antibody incubation: The membrane is incubated with a primary antibody specific to the target protein
6. Washing: Unbound primary antibodies are washed away
7. Secondary antibody incubation: The membrane is incubated with a labeled secondary antibody that binds to the primary antibody
8. Detection: The protein-antibody complex is visualized using various detection methods
Detection Methods
Several detection methods can be used in Western blotting 1 4:
1. Chromogenic detection: Uses enzyme-conjugated secondary antibodies (like HRP or AP) that produce a colored precipitate when exposed to a substrate
2. Chemiluminescent detection: Uses enzyme-conjugated antibodies that produce light when exposed to a substrate, captured on film or by digital imaging systems
3. Fluorescent detection: Uses fluorophore-conjugated antibodies that emit light when excited at specific wavelengths
4. Radioactive detection: Historically used radioactive probes and autoradiography film (less common today)
Applications
Western blotting has numerous applications in research and clinical settings 5 3:
1. Protein identification and characterization: Detecting specific proteins in complex mixtures
2. Post-translational modifications: Identifying modifications like phosphorylation, glycosylation, and ubiquitination
3. Protein expression analysis: Measuring relative protein levels in different samples
4. Clinical diagnostics: Confirming diseases like HIV through antibody detection
5. Drug development: Evaluating protein targets and drug effects
6. Epitope mapping: Identifying antibody binding sites on proteins
7. Anti-doping testing: Detecting prohibited substances in sports
Advantages
1. Requires only small amounts of reagents 5
2. High specificity due to antibody-antigen interactions 4
3. Provides information about protein molecular weight 1
4. Same protein transfer can be used for multiple analyses 5
5. Can detect proteins at picogram levels with optimized protocols
Limitations
1. Requires specific antibodies for target proteins 5
2. Potential for antibody cross-reactivity and off-target effects 5
3. Time-consuming procedure (typically 1-2 days) 1
4. Semi-quantitative rather than fully quantitative 1
5. Relatively costly due to antibody expenses and detection reagents 5
📝 Word Embedding
machine-learning
A specific type of that maps words to vectors of real numbers, capturing semantic relationships between words. Similar words cluster together in the space, allowing models to understand word meanings and relationships. Examples include Word2Vec and GloVe.
🧬 Xenologs
biology
• Definition: Homologous genes acquired through horizontal gene transfer (HGT) between different species rather than through vertical inheritance.
• Key characteristics:
- Phylogenetic incongruence with species tree
- Often have nucleotide composition or codon usage distinct from host genome
- May be flanked by mobile genetic elements
- Frequently confer novel adaptive functions
• Common mechanisms of transfer:
- Transformation: Uptake of environmental DNA
- Conjugation: Direct cell-to-cell transfer
- Transduction: Virus-mediated transfer
- Endosymbiotic gene transfer: From organelles to nucleus
• Prevalence across life:
- Common in prokaryotes (bacteria and archaea)
- Less frequent but significant in unicellular eukaryotes
- Rare but documented in multicellular eukaryotes
- Extensive in certain lineages (e.g., bdelloid rotifers)
• Biological and evolutionary significance:
- Rapid acquisition of adaptive traits (e.g., antibiotic resistance)
- Metabolic innovation and niche expansion
- Acceleration of evolutionary change
- Complication of phylogenetic reconstruction
• Applications:
- Tracking antibiotic resistance spread
- Identifying potential bioremediation genes
- Understanding microbial genome evolution
- Developing novel antimicrobial strategies
• Key characteristics:
- Phylogenetic incongruence with species tree
- Often have nucleotide composition or codon usage distinct from host genome
- May be flanked by mobile genetic elements
- Frequently confer novel adaptive functions
• Common mechanisms of transfer:
- Transformation: Uptake of environmental DNA
- Conjugation: Direct cell-to-cell transfer
- Transduction: Virus-mediated transfer
- Endosymbiotic gene transfer: From organelles to nucleus
• Prevalence across life:
- Common in prokaryotes (bacteria and archaea)
- Less frequent but significant in unicellular eukaryotes
- Rare but documented in multicellular eukaryotes
- Extensive in certain lineages (e.g., bdelloid rotifers)
• Biological and evolutionary significance:
- Rapid acquisition of adaptive traits (e.g., antibiotic resistance)
- Metabolic innovation and niche expansion
- Acceleration of evolutionary change
- Complication of phylogenetic reconstruction
• Applications:
- Tracking antibiotic resistance spread
- Identifying potential bioremediation genes
- Understanding microbial genome evolution
- Developing novel antimicrobial strategies
🧬 Zinc Fingers
biology
• Definition: Protein structural motifs that coordinate one or more zinc ions to stabilize their fold and facilitate interactions with other molecules, particularly DNA, RNA, or proteins.
• Structure and classification:
- Classical C2H2 zinc fingers: Contain two cysteines and two histidines that coordinate a zinc ion
- C4 zinc fingers: Four cysteine residues coordinate the zinc ion (e.g., nuclear hormone receptors)
- C3H zinc fingers: Three cysteines and one histidine coordinate the zinc ion
- RING finger : Cross-brace arrangement of cysteines and histidines coordinating two zinc ions
• Biological functions:
- Transcription regulation: Binding to specific DNA sequences to control gene expression
- RNA binding: Recognizing specific RNA structures or sequences
- Protein-protein interactions: Mediating interactions between proteins in cellular processes
- : Contributing to changes in chromatin structure and accessibility
• Applications in biotechnology:
- Zinc finger nucleases (ZFNs): Engineered proteins combining zinc finger with nuclease activity for genome editing
- Artificial transcription factors: Custom-designed zinc fingers fused to regulatory
- Protein engineering: Creating novel binding specificities for research and therapeutic applications
- Diagnostic tools: Zinc finger-based probes for detecting specific nucleic acid sequences
• Clinical relevance:
- Cancer biology: Mutations in zinc finger proteins associated with various cancers
- Developmental disorders: Defects in zinc finger proteins linked to congenital abnormalities
- Therapeutic targets: Potential for targeting disease-associated zinc finger proteins
- Gene therapy: ZFN-based approaches for correcting genetic mutations
- Circular dichroism: Measuring optical activity
- NMR spectroscopy: Distinguishing based on chemical environment
- Computational methods: Predicting and analyzing stereochemical properties
• Applications in bioinformatics:
- Molecular docking: Accounting for stereochemistry in protein-ligand interactions
- Structure-based drug design: Optimizing stereochemistry for target binding
- Conformational analysis: Predicting energetically favorable
- Cheminformatics: Representing and searching stereochemical information in databases
• Structure and classification:
- Classical C2H2 zinc fingers: Contain two cysteines and two histidines that coordinate a zinc ion
- C4 zinc fingers: Four cysteine residues coordinate the zinc ion (e.g., nuclear hormone receptors)
- C3H zinc fingers: Three cysteines and one histidine coordinate the zinc ion
- RING finger : Cross-brace arrangement of cysteines and histidines coordinating two zinc ions
• Biological functions:
- Transcription regulation: Binding to specific DNA sequences to control gene expression
- RNA binding: Recognizing specific RNA structures or sequences
- Protein-protein interactions: Mediating interactions between proteins in cellular processes
- : Contributing to changes in chromatin structure and accessibility
• Applications in biotechnology:
- Zinc finger nucleases (ZFNs): Engineered proteins combining zinc finger with nuclease activity for genome editing
- Artificial transcription factors: Custom-designed zinc fingers fused to regulatory
- Protein engineering: Creating novel binding specificities for research and therapeutic applications
- Diagnostic tools: Zinc finger-based probes for detecting specific nucleic acid sequences
• Clinical relevance:
- Cancer biology: Mutations in zinc finger proteins associated with various cancers
- Developmental disorders: Defects in zinc finger proteins linked to congenital abnormalities
- Therapeutic targets: Potential for targeting disease-associated zinc finger proteins
- Gene therapy: ZFN-based approaches for correcting genetic mutations
- Circular dichroism: Measuring optical activity
- NMR spectroscopy: Distinguishing based on chemical environment
- Computational methods: Predicting and analyzing stereochemical properties
• Applications in bioinformatics:
- Molecular docking: Accounting for stereochemistry in protein-ligand interactions
- Structure-based drug design: Optimizing stereochemistry for target binding
- Conformational analysis: Predicting energetically favorable
- Cheminformatics: Representing and searching stereochemical information in databases
